Back to Journals » Cancer Management and Research » Volume 10
FOXA1 is upregulated in glioma and promotes proliferation as well as cell cycle through regulation of cyclin D1 expression
Authors Zhang C, Yang M, Li Y, Tang S, Sun X
Received 14 March 2018
Accepted for publication 28 April 2018
Published 6 September 2018 Volume 2018:10 Pages 3283—3293
DOI https://doi.org/10.2147/CMAR.S168217
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Harikrishna Nakshatri
Chuanqiang Zhang,1 Meixia Yang,2 Yanmin Li,1 Suwen Tang,1 Xizhou Sun1
1Department of Neurosurgery, Shandong Rizhao City Hospital of Traditional Chinese Medicine, Rizhao, Shandong, 276800, People’s Republic of China; 2Department of Neurology, Shandong Rizhao City Hospital of Traditional Chinese Medicine, Rizhao, Shandong, 276800, People’s Republic of China
Introduction: Forkhead box A1 (FOXA1) has been found to upregulate in numerous cancers, such as ovarian cancer and glioma. However, the detailed function of FOXA1 in glioma is still not known. The purpose of this study was to explore the underlying mechanism of FOXA1 in glioma cell progression.
Methods: The expressions of FOXA1 in glioma tissues and cells were determined using quantitative reverse transcription-polymerase chain reaction and western blotting assays. Wound healing assay and Transwell invasion assay were employed to detect the effects of FOXA1 on cellular migration and invasion. Cell Counting Kit-8 assay, colony formation assay, and flow cytometry analyses were also performed.
Results: Our study results suggested FOXA1 was upregulated in glioma tissues and cells and revealed that FOXA1 promoted glioma cellular proliferation by facilitating G1/S transition. Previous work has indicated that CCND1 expression is regulated by FOXA1 in ovarian cancer. ChIP and qChIP assay as well as dual luciferase reporter assay validated that CCND1 expression was also regulated by FOXA1 in glioma cells. Moreover, over-expression of CCND1 in siFOXA1-transfected cells partly offsets the effect of FOXA1 inhibition on cellular proliferation.
Conclusion: FOXA1 promotes glioma cell progression, including cell proliferation and cell cycle, by targeting CCND1, and shows potential for the development of targeted treatment for glioma.
Keywords: FOXA1, proliferation, invasion, CCND1, glioma
Introduction
Glioma has become one of the most common neuroectodermal brain cancers in adults, accounting for 50%–55% of the total brain cancers.1,2 Despite the development of different treatment strategies, such as surgical resection followed by radiation and chemotherapy, the prognosis of patients with the most malignant form of glioma still remains at just 12–15 months.3–5 Thus, it is necessary to develop novel and effective therapeutic strategies for glioma.
Forkhead box A1 (FOXA1), a transcription factor, belongs to the forkhead box family.6 FOXA proteins consist of three members, FOXA1, FOXA2 and FOXA3.7 FOXA1 contains the N- and C-terminal transactivation domains and forkhead DNA-binding domain.8 Increasing evidence indicates that FOXA factors are involved in the development of endoderm-derived organs such as the liver, pancreas, lungs, and prostate.9 FOXA1 regulates several gene expressions involved in the development of multiple tissues, including the mammary gland, midbrain, lung, liver as well as progression of several tumors.10,11 Additionally, FOXA1 has been shown to open closed chromatin and facilitates the recruitment of other transcription factors.12 FOXA1 also regulates cyclin D1 expression in ovarian cancer.13 A previous report has indicated that FOXA1 is upregulated in glioma tissues;14 however, the detailed mechanism of FOXA1 is not known.
Cyclin D1 (CCND1) acts as a key regulatory protein that plays a critical role in cell cycle. A previous report has suggested that ectopic expression of cyclin D1 promotes cell cycle.15 Therefore, cyclin D1 is considered as a key regulator of the cell cycle. Abnormal expression of cyclin D1 has also been found in the pathogenesis of lung carcinoma and is associated with poor prognosis.16
In the present study, we determined the expression of FOXA1 in glioblastomas multiforme (GBM) tissues and cell lines. Subsequently, functional gain and loss analyses were performed to determine the effect of FOXA1 on glioma cell proliferation, migration and invasion.
Materials and methods
Cell culture
Normal human astrocytes (NHAs) were obtained from Lonza (Basel, Switzerland) and cultured in astrocyte growth media (Lonza) containing 3% fetal bovine serum (FBS), 4.5% glucose and reagents from BulletKits (Lonza) in a humidified incubator at 37°C and 5% CO2. Human GBM cell lines U87-MG, U251 and HEK-293T were purchased from Chinese Academy of Sciences Cell Bank (Shanghai, People’s Republic of China), and were cultured in DMEM (Hyclone, Logan, UT, USA) supplemented with 100 units of penicillin/mL, 100 ng of streptomycin/mL as well as 10% FBS. All the cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
Tissue samples and ethics statement
A total of 46 pairs of human glioma tissues and 16 adjacent normal tissues were available from the Department of Neurosurgery, Shandong Rizhao City Hospital of Traditional Chinese Medicine. Glioma specimens were verified and classified by three experienced clinical pathologists according to the WHO standard classification of tumors. Tumor tissues were cooled in liquid nitrogen directly from the operating room and stored at −80°C. This study was approved by The Research Ethics Committee of Shandong Rizhao City Hospital of Traditional Chinese Medicine. All patients signed a written informed consent form.
Western blotting analysis
The harvested cells were lysed using RIPA buffer (150 mM NaCl, 1% NP-40, 1 mM ethylenediaminetetraacetic acid, 50 mM Tris-HCl, pH 7.4, 0.25% sodium deoxycholate) and protease inhibitor cocktail (Roche Diagnostics, Mannheim, German). The protein concentration was measured using a BCA protein assay kit (Pierce, Waltham, MA, USA) according to manufacturer’s protocol. Equal amounts of protein were separated electrophoretically on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes (Roche, Basel, Switzerland). This was followed by blocking with 5% skim milk at room temperature for 1 h; the membranes were incubated with indicated antibodies at 4°C overnight. After washing with Phosphate Buffered Solution Tween-20 three times, the membranes were incubated with horseradish peroxidase (HRP)-conjugated second antibodies at room temperature for 1 h. Finally, the blots were visualized using enhanced chemiluminescence reagent (Pierce, Minneapolis, MN, USA) according to manufacturer’s instructions. The antibodies used were as follows: FOXA1 (1:1000; Abcam, Cambridge, UK; ab23738), cyclin D1 (1:3000; Abcam; ab134175), β-actin (1:5000; Abcam; ab8227), HRP-conjugate goat anti-mouse (1:5000; Abcam; ab214879) and HRP-conjugate goat anti-rabbit (1:5000; Abcam; ab214880).
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissues and cells using Trizol (Invitrogen, Shanghai, People’s Republic of China) according to the manufacturer’s instructions. After purification, 2 μg total RNA was utilized to synthesize complementary DNA (cDNA) through a PrimeScript RT reagent Kit gDNAEraser (TaKaRa, Tokyo, Japan). Subsequently, SYBR Green Realtime PCR Master Mix (Roche Diagnostics) was utilized to perform qRT-PCR assay. The cDNA fragments were denatured at 95°C for 20 s, annealed at 57°C for 15 s and extended at 72°C for 40 s for 35 cycles with the ABI PRISM 7500 Quantitative PCR system (Life Technologies, Carlsbad, CA, USA). The PCR products were normalized to GAPDH, and relative expression of mRNA was calculated using 2−DDCT method. Each sample was examined in triplicate. FOXA1 forward, 5′-GCAATACTCGCCTTACGGCT-3′; reverse, 5′-TACACACCTTGGTAGTACGCC-3′; CCND1 forward, 5′-CCTCGGTGTCCTACTTCAAA-3′; and reverse, 5′-GGGATGGTCTCCTTCATCTT-3′; GAPDH forward, 5′-ACCACAGTCCATGCCATCAC-3′; reverse, 5′-TCCACCACCCTGTTGCTGTA-3′.
Wound healing assay
Approximately, 1 × 106 transfected U87-MG or U251 cells were placed in a 6-well plate and cultured to 95–100% confluence. The monolayers were scratched with a 20 μL pipette tip, and the detached cells were removed using cold PBS and cultured in serum-free medium. The wounds were observed under a light microscope and photographed at 0 h and 48 h. The width of wound was measured by ImageJ software (National Institutes of Health, Bethesda, MD, USA). The distance of cell migration was represented by: (width of the original wound (0 h) – width of wound at 48 h)/width of the original wound (0 h) × 100%. Each sample was examined in triplicate.
Transwell invasion assay
Transwell cell culture chambers (BD Bioscience, San Jose, CA, USA) and Matrigel (BD Bioscience, San Jose, CA, USA) were used for the Transwell invasion assay. Briefly, the culture chambers were first coated with 80 μL Matrigel (Matrigel:merum-free medium =1:8) and maintained at 37°C for 30 min. Subsequently, approximately 5 × 104 transfected cells were resuspended in 200 μL serum-free medium and placed in the upper compartment of the Transwell inserts. The bottom chambers contained 500 μL of complete medium. After incubation at 37°C for 36 h, cells on the upper chamber were removed using a cotton swab, and the invaded cells at the bottom of the top chamber were stained with 0.1% crystal violet at room temperature for 15 min, and counted as well as photographed under an Olympus fluorescence microscope (Tokyo, Japan). Each sample was examined in triplicate.
Cell Counting Kit-8 (CCK-8) assay
CCK-8 assay was used to determine the effect of FOXA1 on cellular proliferation. In brief, 5 × 103 transfected U87-MG or U251 cells were seeded in 96-well plates with 200 μL complete medium. Twenty microliters of CCK-8 reagents was added at 0 h, 24 h, 48 h and 72 h. After 30 min of incubation with CCK-8 reagent at 37°C, the absorbance at 450 nm was measured and used for calculating cell viability. Each sample was examined in triplicate.
Colony formation assay
Colony formation assay was utilized to identify the effect of FOXA1 on cell proliferation. Briefly, 5 × 103 transfected U87-MG or U251 cells were seeded in a new 6-well plate and cultured for approximately 14 days. Colonies were fixed with methanol at room temperature for 10 min and stained with 0.1% crystal violet (Sigma-Aldrich Co., St Louis, MO, USA) at room temperature for 10 min. The colonies (cells > 50) were counted as well as photographed under a light microscope. Each sample was examined in triplicate.
Cell cycle analysis
After transfection for 48 h, cells were harvested by centrifugation at 500× g for 5 min. After washing with cold PBS three times, cells were fixed with 75% alcohol and stored at −20°C overnight. Subsequently, cells were washed with cold PBS three times before adding 1 mL PBS containing 40 μg propidium iodide and 100 μg RNase A. Lastly, the cells were analyzed by flow cytometry. Each sample was examined in triplicate.
ChIP and qChIP assay
Chromatin Immunoprecipitation Assay kit (Beyotime, Jiangsu, People’s Republic of China) was utilized to perform ChIP analysis according to manufacturer’s instructions. In brief, U87-MG and U251 cells were grown to 90–100% confluence. Subsequently, cells were washed with cold PBS three times and chemically cross-linked with 1% formaldehyde for 20 min at 37°C. Next, cells were lysed with lysis buffer at 4°C for 1 h and sonicated three cycles at 4°C, each cycle for 15 times. Three micrograms of anti-rabbit IgG and FOXA1 antibody were added to the lysis solution and incubated at 4°C overnight. Protein A beads were used to isolate FOXA1- or IgG-interacted DNA fragments. PCR Purification kit (Qiagen, Shanghai, People’s Republic of China) was used to purify the binding chromatin. Each experiment was performed at least three times. The sequence of CCND1 used was as follows: forward, 5′-GTGGCAGGCTTGGCGGATGT-3′; reverse, 5′-TTGGTTGTCACGGCGGGTGG-3′.
Dual luciferase reporter assay
The promoter region (−2000 to +200) of CCND1 was amplified and cloned into pGL3 control vector (Invitrogen). HEK-293T cells were placed in a 24-well plate and co-transfected with CCND1 luciferase reporter, Renilla and vector or FOXA1 (0.5 μg, 1 μg and 2 μg). After transfection for 24 h, the luciferase activity was determined using the Promega Dual Luciferase Reporter Assay System (Madison, WI, USA) according to manufacturer’s instructions. Each sample was examined in triplicate.
Statistical analysis
Analysis was done with the SPSS software version 18.0 for Windows (SPSS, Chicago, IL, USA). All data are represented as means ± standard deviation. The statistical differences between multiple groups were determined by one-way analysis of variance (ANOVA) and Tukey test. Two groups were compared using an unpaired, two-tailed Student’s t-test. Survival probability was calculated using the Kaplan-Meier method followed by log-rank test. Statistical significance was identified as p < 0.05 for all comparisons.
Results
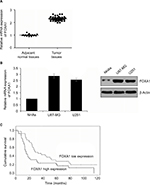
FOXA1 expression is upregulated in glioma tissues and cells
A previous report has indicated that FOXA1 is upregulated in glioma tissues,14 but the detailed mechanism of FOXA1 in glioma has not been known. Here, we first collected 46 glioma tissue samples and 16 adjacent normal tissue samples, and detected the expression of FOXA1 using qRT-PCR. As shown in Figure 1A, the expression of FOXA1 in tumor tissues was higher than that in normal tissues (Figure 1A). Subsequently, we also detected FOXA1 expression in glioma cells, compared with normal glial cells. Results of qRT-PCR and western blotting validated the upregulation of FOXA1 in glioma cells (Figure 1B). Additionally, we analyzed the correlation of FOXA1 expression and clinicopathological features of glioma. In 46 glioma tissues, high FOXA1 expression was markedly more common in glioma tissues with high-grade glioma than in those with low-grade glioma (Table 1, p < 0.05). Additionally, high expression of FOXA1 was associated with tumor size and neck lymph node metastasis (Table 1, p < 0.05). However, there was not any statistically significant correlation of FOXA1 expression with age or sex (Table 1, p > 0.05). Moreover, prominent expression of FOXA1 predicted a poor prognosis of GBM patients (Figure 1C, p < 0.05).
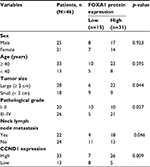  | Table 1 Clinicopathological variables in 46 GBM patients Abbreviations: FOXA1, forkhead box A1; GBM, glioblastomas multiforme. |
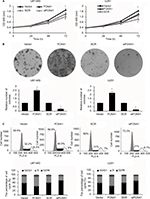
Upregulation of FOXA1 facilitates migration and invasive ability of glioma cells
To further decipher the function of FOXA1 in glioma development, we over-expressed or knocked down FOXA1 in glioma cell lines, including U87-MG and U251. The expression of FOXA1 was detected using qRT-PCR and western blotting assay (Figure 2A and B). Wound healing assay and transwell invasion assay were performed to determine the effect of FOXA1 on cell migration and invasion, respectively. The result of the wound healing assay suggested markedly higher migration rate in FOXA1 overexpression group and lower migration rate in FOXA1 inhibition group in comparison with each individual control group (Figure 2C, p < 0.05). Results of the transwell assay indicated that more cells penetrated across the membrane in the FOXA1 over-expression group and there were a lot fewer invaded cells in the FOXA1 inhibition group, compared with each individual control group (Figure 2D, p < 0.05). The abovementioned results indicate that upregulation of FOXA1 facilitates migration and invasive ability of glioma cells.
Ectopic expression of FOXA1 promotes cell proliferation of glioma cells through regulation of cell cycle
Subsequently, we assessed whether FOXA1 regulated cell proliferation of glioma cells. CCK-8 and colony formation assay were performed to determine the effect of FOXA1 on cell proliferation. As shown in Figure 3A, ectopic expression of FOXA1 significantly increased the rate of cell proliferation in U87-MG and U251 cells, and inhibition of FOXA1 decreased the rate of cell proliferation in U87-MG and U251 cells (Figure 3A). Similar results were observed in the colony formation assay. The number of colonies was increased 2.5-fold in FOXA1-transfected cells, compared with the control group. Meanwhile, inhibition of FOXA1 reduced the number of colonies (Figure 3B). Because cell proliferation is directly connected to cell cycle distribution, we hypothesized as to whether FOXA1 promoted the proliferation of glioma cells via regulation of cell cycle. Flow cytometry analysis demonstrated that ectopic expression of FOXA1 decreased the percentage of G0/G1 phase cells and increased the percentage of S and G2/M phase cells (Figure 3C). However, inhibition of FOXA1 increased the percentage of G0/G1 phase cells and decreased the percentage of S and G2/M phase cells. The abovementioned results indicate FOXA1 promotes the proliferation of U87-MG and U251 cells through regulation of cell cycle.
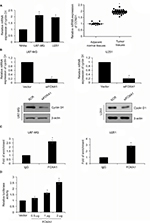
CCND1 is a direct target of FOXA1 in glioma cells
Cell cycle is regulated by the cyclin family, cyclin-dependent kinase (CDK) and CKI. Cyclin family consists of CCNA-Y, CCNA2, CCNB1, CCND1 as well as CCNE1.17,18 We first detected the expression of CCNDA1 in glioma cells and tissue samples, when compared with normal glial cells or adjacent normal tissues, respectively. Results of qRT-PCR and western blotting validated the upregulation of FOXA1 in glioma cells (Figure 4A). Previous work has found that FOXA1 regulated CCND1 expression in ovarian cancer.13 To further determine whether FOXA1 promoted cell cycle in glioma through regulation of CCND1 expression, we measured the CCND1 expression in glioma specimens. The results revealed that the expression of CCND1 was positively associated with expression of FOXA1 (Table 1, p < 0.05). Subsequently, we detected whether FOXA1 regulated CCND1 expression using qRT-PCR and western blotting assay. As shown in Figure 4A, we found inhibition of FOXA1 reduced the expression of CCND1 (Figure 4B). To further determine whether FOXA1 could directly bind to the promoter region of CCND1, we performed ChIP and qChIP assay as well as dual luciferase reporter assay. The result of the ChIP and qChIP assay suggested that compared to IgG group, the fold of enrichment increased almost 2.5-fold, indicating FOXA1 could directly bind to the promoter region of CCND1 (Figure 4C). Luciferase activity in HEK-293T cells was notably increased after FOXA1 was over-expressed in cells (Figure 4D). The abovementioned finding suggests that FOXA1 directly regulates CCND1 expression in glioma cells.
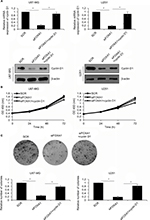
Over-expression of CCND1 partially attenuates FOXA1-depletion inhibited cell growth
Our present work indicated that CCND1 served as a downstream target of FOXA1; so, we further explored the role of FOXA1 in glioma through mediation of CCND1. PcDNA3.1-CCND1 plasmid was transfected into siFOXA1-transfected U87-MG and U251 cells to reverse the reduced CCND1 levels. After transfection, the expression of cyclin D1 was determined by qRT-PCR and western blotting analyses. As illustrated in Figure 5A, the mRNA and protein levels of CCND1 were higher in the siFOXA1+CCND1 group than those in the siFOXA1 group (Figure 5A). To further confirm whether FOXA1 regulated cell proliferation via regulation of CCND1, CCK-8 assay and colony formation analyses were carried out. The results demonstrated that restoration of CCND1 partially rescued FOXA1 inhibition suppressive functions in U87-MG and U251 cells (Figure 5B and C). Collectively, these findings suggested that CCND1 acted as a downstream effector of FOXA1 in the regulation of the proliferation of glioma cells.
Discussion
The transcription factor FOXA1 binds to genomic DNA through the forkhead DNA-binding domain and mediates expression of various genes,8 such as CDH1.19 A previous report has indicated that FOXA1 is upregulated in glioma tissues,14 but the detailed mechanism of FOXA1 in glioma has not been known.
In our study, we found that the expression of FOXA1 in glioma tissues and cells was higher than adjacent normal brain tissue or NHAs. In addition, the FOXA1 expression was positively related to glioma grade, and predicted a poor prognosis. Subsequently, CCK-8 assay and colony formation assay showed that the proliferative capacity of glioma cells was markedly increased or reduced in FOXA1- or siFOXA1-transfected cells. Meanwhile, through flow cytometry analysis, we found that ectopic expression of FOXA1 promotes G1/S transition. As is well known, cell cycle is regulated by protein kinase complex, which is composed of multiple cell cycle proteins (cyclins) and the corresponding CDKs.20,21 A recent study showed that FOXA1 regulated cyclin D1 expression in ovarian cancer.13 In order to verify whether FOXA1 regulated cyclin D1 in glioma cells, we next performed qRT-PCR and western blotting analyses, which suggested that both mRNA and protein levels of cyclin D1 were regulated by FOXA1. Additionally, ChIP and qChIP assay as well as luciferase reporter assay indicated that FOXA1 transcriptionally activated CCND1. Therefore, we suggest that FOXA1 can promote G1/S transition by facilitating the expression of cyclin D1. Additionally, FOXA1 also facilitates glioma cell migration and invasion.
In summary, this study investigated the detailed functions of FOXA1 in the growth and metastasis of glioma cells, and suggests that FOXA1 acts as a novel oncogene in glioma. FOXA1 promotes proliferation as well as cell cycle of glioma cells via targeting cyclin D1. Nevertheless, we need further studies to verify whether FOXA1 promotes glioma cells proliferation and metastasis in vivo.
Disclosure
The authors report no conflicts of interest in this work.
References
Cai J, Zhu P, Zhang C, et al. Detection of ATRX and IDH1-R132H immunohistochemistry in the progression of 211 paired gliomas. Oncotarget. 2016;7(13):16384–16395. | ||
Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005-2009. Neuro Oncol. 2012;14 Suppl 5:v1–49. | ||
Stupp R, Mason WP, van den Bent MJ, et al; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. | ||
Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166–193. | ||
Li QJ, Cai JQ, Liu CY. Evolving molecular genetics of glioblastoma. Chin Med J (Engl). 2016;129(4):464–471. | ||
Jozwik KM, Carroll JS. Pioneer factors in hormone-dependent cancers. Nat Rev Cancer. 2012;12(6):381–385. | ||
Bochkis IM, Rubins NE, White P, Furth EE, Friedman JR, Kaestner KH. Hepatocyte-specific ablation of Foxa2 alters bile acid homeostasis and results in endoplasmic reticulum stress. Nat Med. 2008;14(8):828–836. | ||
Bernardo GM, Keri RA. FOXA1: a transcription factor with parallel functions in development and cancer. Biosci Rep. 2012;32(2):113–130. | ||
Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63(19–20):2317–2328. | ||
Kaestner KH. The FoxA factors in organogenesis and differentiation. Curr Opin Genet Dev. 2010;20(5):527–532. | ||
Augello MA, Hickey TE, Knudsen KE. FOXA1: master of steroid receptor function in cancer. EMBO J. 2011;30(19):3885–3894. | ||
Carroll JS, Liu XS, Brodsky AS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122(1):33–43. | ||
Wang LL, Xiu YL, Chen X, et al. The transcription factor FOXA1 induces epithelial ovarian cancer tumorigenesis and progression. Tumour Biol. 2017;39(5):1010428317706210. | ||
Wang L, Qin H, Li L, et al. Forkhead-box A1 transcription factor is a novel adverse prognosis marker in human glioma. J Clin Neurosci. 2013;20(5):654–658. | ||
Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25(11):1620–1628. | ||
Brennan P, Hainaut P, Boffetta P. Genetics of lung-cancer susceptibility. Lancet Oncol. 2011;12(4):399–408. | ||
Fu XJ, Li HX, Yang K, Chen D, Tang H. The important tumor suppressor role of PER1 in regulating the cyclin-CDK-CKI network in SCC15 human oral squamous cell carcinoma cells. Onco Targets Ther. 2016;9:2237–2245. | ||
Lee B, Sandhu S, McArthur G. Cell cycle control as a promising target in melanoma. Curr Opin Oncol. 2015;27(2):141–150. | ||
Liu YN, Lee WW, Wang CY, Chao TH, Chen Y, Chen JH. Regulatory mechanisms controlling human E-cadherin gene expression. Oncogene. 2005;24(56):8277–8290. | ||
Cheney IW, Neuteboom ST, Vaillancourt MT, Ramachandra M, Bookstein R. Adenovirus-mediated gene transfer of MMAC1/PTEN to glioblastoma cells inhibits S phase entry by the recruitment of p27Kip1 into cyclin E/CDK2 complexes. Cancer Res. 1999;59(10):2318–2323. | ||
Kaldis P, Aleem E. Cell cycle sibling rivalry: Cdc2 vs. Cdk2. Cell cycle. 2005;4(11):1491–1494. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.