Back to Journals » Infection and Drug Resistance » Volume 12
First Reported Nosocomial Outbreak Of NDM-5-Producing Klebsiella pneumoniae In A Neonatal Unit In China
Authors Kong Z, Cai R, Cheng C, Zhang C, Kang H, Ma P, Gu B
Received 10 June 2019
Accepted for publication 20 September 2019
Published 15 November 2019 Volume 2019:12 Pages 3557—3566
DOI https://doi.org/10.2147/IDR.S218945
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Ziyan Kong,1,* Rui Cai,1,* Chen Cheng,1 Chuanling Zhang,2 Haiquan Kang,3 Ping Ma,3 Bing Gu1,3
1Medical Technology School, Xuzhou Medical University, Xuzhou, Jiangsu Province, People’s Republic of China; 2Department of Laboratory Medicine, XuZhou Children’s Hospital, Xuzhou, Jiangsu Province, People’s Republic of China; 3Department of Laboratory Medicine, Affiliated Hospital of Xuzhou Medical University, Xuzhou, Jiangsu Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Bing Gu
Department of Laboratory Medicine, Affiliated Hospital of Xuzhou Medical University, Xuzhou 221006, People’s Republic of China
Tel +8615862163560
Email [email protected]
Purpose: Carbapenem-resistant Klebsiella pneumoniae (CRKP) have emerged worldwide and also being a major threat to children and neonate. In this study, we describe a nosocomial outbreak of NDM-5-producing Klebsiella pneumoniae in neonatal unit of a teaching hospital in China from September 2015 to September 2016.
Patients and methods: We collected 12 carbapenem-resistant K. pneumoniae outbreak strains from 12 newborns and characterized these isolates for their antimicrobial susceptibility, clone relationships, and multi-locus sequence types using vitek-2 compact system, pulsed-field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST). Resistant genes were detected by using PCR and sequencing. Plasmid conjugation experiment was carried out to determine the transferability of carbapenem resistance. PCR-based replicon typing (PBRT), S1 nuclease-PFGE, and southern blotting were conducted for plasmid profiling.
Results: All 12 K. pneumoniae isolates were resistant to carbapenems and carried blaNDM-5, blaTEM-1 and blaSHV-11. Furthermore, PFGE analysis showed that NDM-5-producing K. pneumoniae were clonally related and MLST assigned them to sequence type 337. Conjugative assays showed that plasmids harboring blaNDM-5 gene were self-transmissible. Plasmid analysis suggested that all blaNDM-5 gene located on a ∼45 kb IncX3 type plasmid.
Conclusion: To the best of our knowledge, this is the first report of a clone outbreak of blaNDM-5-carrying K. pneumoniae isolates from neonates. There is an urgent need for effective infection control measures to prevent blaNDM-5 variants from becoming epidemic in the neonates in the future.
Keywords: Klebsiella pneumoniae, carbapenemases, blaNDM-5, ST337, neonate, IncX3
Introduction
The continuous emergence of Carbapenem-resistant Klebsiella pneumoniae (CRKP) has become a serious public-health problem worldwide.1 Among the newly emerging carbapenemases, New Delhi metallo-β-lactamase (NDM) has been considered as a major clinical challenge due to its ability to hydrolyzing all β-lactams antibiotics except monobactams and its rapid spread worldwide.2 Since NDM-1 was first identified in a K. pneumoniae isolate from a Swedish patient who had traveled to New Delhi in 2008,3 21 new blaNDM alleles have been discovered in global.4 The rapid evolution of blaNDM genes represents a huge challenge for clinical infection treatment. In 2011, NDM-5 was first identified in a multidrug-resistant Escherichia coli isolate in the UK.5 The NDM-5 enzyme differs from the prevalent NDM-1 at the amino acid positions, which appears to confer elevated resistance to carbapenems and extended-spectrum cephalosporins. Unlike the prevalent NDM-1 enzyme, the identification of NDM-5 enzymes is not common. Among the Enterobacteriaceae, reports of blaNDM-5-harboring K. pneumoniae isolates are sporadic.6–12 In this study, we investigate an outbreak of blaNDM-5-positive K. pneumoniae strains in neonatal unit of a teaching hospital in China with the aim of determining the molecular basis of the emerging ST337 carbapenem-resistant K. pneumoniae isolates responsible for this outbreak. As far as we know, this is the first description of NDM-5-producing K. pneumoniae nosocomial outbreak in the neonatal ward.
Materials And Methods
Collection Of Clinical Isolates And Patients
The study was conducted in the Department of Neonatology at a teaching hospital in Jiangsu Province, China. There are 120 beds in the Department of Neonatology, with an estimated population of 5000 patient visits per year. From September 2015 to September 2016, 12 non-duplicated strains of carbapenem-resistant K. pneumoniae were isolated from inpatients in newborn medicine ward. Strain identification was performed by Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS; Bruker Daltonik GmbH, Bremen, Germany). In vitro antimicrobial susceptibility testing of isolates was analyzed with VITEK-2 compact system (bioMérieux, Marcy-l’Étoile, France). The electronic medical records of the newborns including patient demographics, neonatal birth information, antimicrobial treatment, and clinical outcomes were retrospectively reviewed.
Antimicrobial Susceptibility Testing
The minimum inhibitory concentrations (MICs) of meropenem (MEM), imipenem (IPM), aztreonam (ATM), ceftazidime (CAZ), cefepime (FEP), cefotaxime (CTX), amoxicillin-clavulanate (AMC), sulbactam/cefoperazone (SCF), piperacillin-tazobactam (TZP), amikacin (AM), levofloxacin (LE), ciprofloxacin (CPFX), compound Sulfamethoxazole (SMZ-TMP), tigecycline (TG) and polymyxin B (PB) were measured by broth microdilution method. Antimicrobial susceptibility testing was interpreted in accordance with the Clinical and Laboratory Standards Institute (CLSI),13 except for sulbactam/cefoperazone, tigecycline and colistin, which were interpreted based on the European Committee for Antimicrobial Susceptibility Testing (EUCAST) criteria.14
Polymerase Chain Reaction (PCR) Amplifications And Sequencing
PCR using the primers mentioned in Table 1 allowed the detection of carbapenemase genes (blaKPC, blaNDM, blaVIM, blaOXA-48 and blaIMP),15–19 common extended-spectrum β-lactamase (ESBL) genes (blaCTX-M-1group, blaCTX-M-2group, blaCTX-M-8 group, blaCTX-M-9 group, blaSHV and blaTEM) and AmpC genes (blaCIT, blaMOX, blaDHA, blaEBC, and blaFOX).20,21 DNA templates were prepared by alkaline lysis method using the kit (MoBio, USA). Amplifications were carried out in 50 µL final volume with 2 µL DNA, 25 µL 2X Taq Master Mix (TaKaRa, Dalian, China), and 2 µM of each primer (Shanghai Sangon, Shanghai, China). PCR conditions for all genes consisted of one cycle at 95°C for 5 mins, followed by 30 cycles at 94°C for 1 min, 72°C for 1 min, and one cycle at 72°C for 5 mins; the annealing step varied depending on the target gene as follows: 55°C/1 min for blaIMP; 59°C/1 min for blaKPC, blaVIM, blaCTX-M-2group, blaMOX; 58°C/1 min for blaNDM, blaCTX-M-8group, blaTEM; 62°C/1 min for blaOXA−48, blaCTX-M-1group, blaDHA, blaFOX; 64°C/1 min for blaCTX-M-9group and blaEBC; 66°C/1 min for blaCIT; 68°C/1 min for blaSHV. Amplified genes were screened by electrophoresis on a 1.5% agarose gel. The positive amplicons were subjected to direct sequencing, and the sequences obtained were compared to reported sequences from GenBank (www.ncbi.nlm.nih.gov/GenBank) using BLAST searches.
 |
Table 1 Primers Used For Amplification Of The Resistance Genes Of K. pneumoniae In This Study |
Molecular Typing
The clonal relationships of the NDM-5 producing K. pneumoniae isolates were analyzed using Pulsed-field gel electrophoresis (PFGE) and whole genomic DNA was digested with XbaI restriction endonuclease (TaKaRa, Dalian, China). Salmonella enterica serotype Braenderup H9812 was used as a marker. The PFGE patterns were compared using BioNumerics software version 5.10, with a cutoff at 90% similarity to indicate identical PFGE types (pulsotypes). Multi-locus sequence typing was also performed for genotyping. Seven housekeeping genes (infB, pgi, mdh, phoE, gapA, tonB and rpoB) were amplified by using primer sequences described on the MLST website.22 Sequence types (STs) were assigned using the MLST database (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html).
Conjugation Assay
A conjugation assay was performed between the isolates harboring blaNDM-5 as donors and E. coli 600 (rifampicin-resistant strain) as a recipient.23 Transconjugants were selected on Brain Heart Infusion agar containing 600 µg/mL rifampicin and 2 µg/mL imipenem for 24 hrs at 35°C and the presence of the blaNDM-5 gene was confirmed by PCR sequencing. Susceptibility testing of the transconjugants was performed as well.
Plasmid Analysis
Plasmid incompatibility types of the transconjugants were identified by PCR-based replicon typing as reported previously.24,25 S1-PFGE and Southern blotting were performed to isolate and localize the resistant plasmid carrying blaNDM-5 gene. Briefly, blaNDM-5 positive isolates embedded in gold agarose gel plugs were digested with S1 nuclease (TaKaRa Biotechnology, Dalian, China) and plasmid DNA was electrophoresed on a CHEF-mapper XA (PFGE) system (Bio-Rad, USA). Subsequently, the DNA fragments were blotted onto a positive-charged nylon membrane (Millipore, USA) and hybridized with blaNDM-5 specific probe according to the protocol of DIG High Prime DNA Labeling and Detection Starter Kit I (Roche Applied Sciences, Penzberg, Germany).
Results
Clinical Characteristics Of 12 K. pneumoniae
In September 2015, a premature baby was born in this hospital with neonatal pneumonia and neonatal respiratory distress syndrome (case 1). The first carbapenem-resistant K. pneumoniae strain was isolated from the sputum culture on September 10, 2015. The first baby’s mother suffered from acute gastroenteritis in the 7th month of pregnancy and hospitalized in another hospital in Jiangsu. After 1 day, another neonate also had neonatal respiratory distress syndrome and was positive for carbapenem-resistant K. pneumoniae in sputum culture. No other isolates were detected between October and November. However, since December 2015, the isolation rate of the strain has increased significantly, and 10 strains have been isolated from December 2015 to September 2016. By September 2016, 12 cases had occurred in newborn medicine ward in total. All the 12 neonates were improved after receiving various antimicrobial therapies including moxalactam, ceftazidime, cefotaxime, amoxicillin-sulbactam, amoxicillin-clavulanate and piperacillin-tazobactam. The 12 NDM-5-positive K. pneumoniae strains were detected from urine (n=1), blood (n=2) and sputum (n=9), respectively. Eleven newborns had neonatal pneumonia, three had neonatal respiratory distress syndrome, and two neonatal sepsis. Four newborns were premature, one of whom were very low birth weight infant. The demographic and clinical profiles of the 12 newborns involved in the outbreak of NDM-5-positive K. pneumoniae isolates are shown in Table 2.
 |
Table 2 Clinical Features Of The Neonates In This Study |
Antibiotic Susceptibility Testing And Detection Of Resistance Determinants
All 12 clinical isolates exhibited resistance to meropenem, imipenem, aztreonam, cefotaxime, ceftazidime, cefepime, sulbactam-cefoperazone, amoxicillin-clavulanate and piperacillin-tazobactam but remained susceptible to levofloxacin, ciprofloxacin, amikacin, compound sulfamethoxazole, tigecycline and polymixin B. All 12 isolates carried the blaNDM-5, blaSHV-11, and blaTEM-1 genes. The other resistance genes present in clinical K. pneumoniae isolates were not detected. The antibiotic resistance characteristics of the 12 NDM-5-positive K. pneumoniae isolates and their corresponding transconjugants are shown in Table 3.
 |
Table 3 Antibiotic Resistance Characteristics Of The 12 NDM-5-Positive K. pneumoniae Isolates And Their Corresponding Transconjugants |
Molecular Typing
PFGE typing revealed that all 12 K. pneumoniae isolates in this outbreak shared highly similar PFGE patterns, which suggests that the 12 isolates were clonally related (Figure 1). MLST analysis showed that these 12 strains, KP1–12, belonged to ST337 with the allelic profile 2-1-11-1-1-1-13.
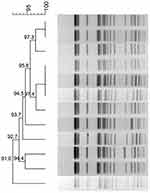 |
Figure 1 Dendrogram of PFGE profiles of 12 blaNDM-5-positive Klebsiella pneumoniae isolates. |
Conjugation And Plasmid Analysis
In our study, all of plasmids carried blaNDM-5 gene from the donor carbapenem-resistant K pneumoniae strains were successfully transferred to the recipient Escherichia coli EC600. The corresponding transconjugants are termed EC600-1–EC600-12, respectively. The MIC values of 12 the transconjugants were tested, and all transconjugants exhibited resistance to carbapenem and enzyme inhibitors (Table 3). As shown in Figure 2, S1-PFGE and Southern blot hybridization using a DIG-labeled blaNDM−5 with incompatibility group-specific probe revealed that blaNDM-5 was located on the nearly same size (~45 kb) IncX3 plasmids.
Discussion
Carbapenem-resistant Enterobacteriaceae are an emerging problem, which spreads among our most vulnerable populations, children.26 Many carbapenem resistance genes, such as blaKPC-2,21 blaNDM-1,27 blaOXA-48,28 blaOXA-232 and blaIMP-38,29,30 have been found in K. pneumoniae outbreak strains in the newborn. Although NDM-1 enzyme was previously reported as the most prevalent type of carbapenemase in children,31 our current results suggested that the emergence of NDM-5-producing K. pneumoniae caused nosocomial outbreak in neonatal unit. Our research team previously found high prevalence of blaNDM-5 variants among carbapenem-resistant E. coli in Northern Jiangsu Province.32 Based on analysis of previous articles, this is the first identification of blaNDM-5-harboring K pneumoniae outbreak strains in the neonatal infection. In this study, we aimed to identify the microbial resistance characteristics and transmission mechanism of CPKP infections caused by NDM-5-producing K. pneumoniae isolates, which may help to prevent the blaNDM-5 from becoming epidemic in neonates.
Previous study found that premature birth and very low birth weight are associated with neonatal nosocomial infections with carbapenem-resistant Enterobacteriaceae.33 In this study, 33.3% and 8.3% of the 12 newborns had premature birth and very low birth weight, respectively. During the study period, 7.0% and 3.15% of all patients in the neonatal unit had premature birth and very low birth weight, respectively. Newborns at high risk should be of the greatest concern in the future when performing procedures to prevent CRKP infection. Compared to adults, children have more limited therapeutic options.34 In our study, all the clinical strains were susceptible to tigecycline and colistin in vitro. However, tigecycline is not recommended for children due to the risk of dental staining and colistin is not currently used in clinical treatment of children in China. Besides, aminoglycosides and fluoroquinolones are also rarely used for children due to their nephrotoxicity and ototoxicity.35 Clinical data show that all the 12 neonates were improved after receiving various antimicrobial therapies including moxalactam, ceftazidime, cefotaxime, amoxicillin, sulbactam, amoxicillin-clavulanate and piperacillin-tazobactam. In this study, 12 newborns responded to treatment with these agents, which seems inconsistent with the antibiotic resistance characteristics of the clinical isolates. We assumed when these agents are combined, synergistic bactericidal action against pathogens may be achieved in vivo.36 The isolates detected from sputum samples were presumed to be the flora colonized in the respiratory tract rather than the real infectious pathogen.
PFGE analysis showed that 12 clinical strains shared highly similar PFGE patterns in this outbreak. This implied that the outbreak of NDM-5-producing K. pneumoniae was because of clone spread and the first newborn might be the source of this outbreak. The first baby’s mother was hospitalized for acute gastroenteritis during pregnancy and had high-risk factors for intrauterine infection. Although the NDM-5-producing K. pneumoniae strain was not detected from the mother, we assumed that a small number of these strains might potentially have been colonized in the mother’s body and infected the fetus through the fetal circulation. Then, the CRKP isolate spread among patients in the neonatal ward, more research would be needed to uncover it in the future. It is noteworthy that blaNDM-5 were not only reported in clinical specimens but also in hospital environmental such as the incubator water and the sharing of breast milk.37,38 Hence, we cannot deny that the hospital environment may be the diffusion reservoirs of NDM-5-producing bacteria. Regrettably, the origin of the NDM-5-producing strains in our study is still unknown. Personal contact between the caregivers and the newborns hospitalized in the same ward is the most likely route of the transmission of the isolates. Hospital infection department took strict measures to control the outbreak in the hospital. First, pre-screening of carbapenem-resistant Enterobacteriaceae(CRE) in sputum samples and rectal swabs were introduced before admission to the neonatal medical ward. Second, strict isolation procedures were implemented for patients with CRE infection. Third, it was necessary for medical staff who contact with patients infected with CRE to go through a disinfection procedure. Finally, the neonatal medical wards where newborns with CRE infection stayed were thoroughly sterilized after the discharge of the patients. The sterilized ward left unoccupied for more than 2 weeks before new patients were admitted. Since the implementation of these strict measures in this neonatal medical ward, no infections due to ST337 NDM-5-producing K. pneumoniae isolates have occurred as of May 2017.
Several countries, such as India (ST648),39 Denmark (ST1284),40 Poland (ST418),41 Spain (ST418),42 Japan (ST540),43 and Singapore (ST231),10 have reported infections caused by NDM-5-positive strains since the initial identification of this carbapenemase type in UK (ST648).5 In China, ST167 carbapenemase-producing Escherichia coli is main reservoir of blaNDM-5.44 This is the first identification of NDM-5-producing K. pneumoniae isolate belonged to ST337, which related to CC37, and have only been identified in KPC-2-producing K. pneumoniae in carbapenem-resistant Enterobacteriaceae family.
The conjugation experiment showed that the carbapenem resistance phenotype of the 12 clinical isolates could be successfully transferred to the recipient strain EC600. All 12 transconjugants only carried the blaNDM-5, and the blaSHV-11 and blaTEM-1 were not detected. The plasmid analysis showed that all blaNDM-5 were located on the IncX3 plasmids of the same size (with approximate size 45 kb). Notably, IncX3-type plasmids of 45 kb carrying blaNDM-5 genes have been identified in India,6 Australia and Denmark.38,39 Furthermore, previous studies revealed that the IncX3-type plasmids played a significant role in the dissemination of the blaNDM-5 gene among Enterobacteriaceae in China.45 Therefore, there is an urgent need to adopt effective measures to control the spread of this resistant plasmid type.
Although international travel previously reported contributed to the global spread of NDM enzyme,3 all patients in our study who carry blaNDM-5 gene have never been abroad. It was assumed that these NDM-5-producing isolates were autochthonous clones that had obtained the plasmid carrying blaNDM-5.44 Notably, recent studies in China have revealed that NDM-5-producing Enterobacteriaceae was identified from pigs, dairy cows, and vegetables.46–48 And, animals and plants were already important sources of drug-resistant strains. There is an urgent need for more epidemiological studies in the future to better understand mechanisms of emergence and dissemination of blaNDM-5 gene in China.
Conclusion
Our study first described the outbreak of ST337 NDM-5-producing K. pneumoniae isolates from neonates in China. Moreover, these outbreak strains were also detected to carry blaSHV-11 and blaTEM-1. Their resistance profile may help pediatricians promote the prudent use of antibiotics in child health care. Worldwide surveillance of these ST337 NDM-5 producing K. pneumoniae isolates from neonates and implementation of stricter control measures are urgently needed to prevent these multiple-resistant strains from further disseminating in neonatal ward.
Ethical Approval
Clinical research ethics committee of the Affiliated Hospital of Xuzhou Medical University approved the study as all samples collected in this work were initially used to diagnose for patient care without increasing the patient’s medical costs and suffering.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (81871734, 81471994), Jiangsu Provincial Natural Science Foundation (BK20151154), Jiangsu Provincial Medical Talent (ZDRCA2016053), Six Talent Peaks Project of Jiangsu Province (WSN-135), and Advanced Health Talent of Six-One Project of Jiangsu Province (LGY2016042).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Tzouvelekis LS, Markogiannakis A, Psichogiou M, et al. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev. 2012;25(4):682–707.
2. Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10(9):597–602. doi:10.1016/S1473-3099(10)70143-2
3. Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-β-Lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–5054. doi:10.1128/AAC.00774-09
4. Liu L, Feng Y, McNally A, Zong Z. BlaNDM-21, a new variant of blaNDM in an Escherichia coli clinical isolate carrying blaCTX-M-55 and rmtB. J Antimicrob Chemother. 2018;73(9):2336–2339. doi:10.1093/jac/dky226
5. Hornsey M, Phee L, Wareham DW, Novel Variant A. NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents & Chemother. 2011;55(12):5952–5954. doi:10.1128/AAC.05108-11
6. Krishnaraju M, Kamatchi C, Jha AK, et al. Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J Med Microbiol. 2015;33(1):30–38. doi:10.4103/0255-0857.148373
7. Liu P, Liu Y, Wang L, Wei -D-D, Wan L-G. Draft genome sequence of an NDM-5-producing Klebsiella pneumoniae sequence type 14 strain of serotype K2. Genome Announc. 2016;4(2):e01610–e01615. doi:10.1128/genomeA.01610-15
8. Du H, Chen L, Tang Y, Kreiswirth BN. Correspondence emergence of the mcr-1 colistin resistance gene in carbapenem-resistant. Lancet Infect Dis. 2016;16(3):287–288. doi:10.1016/S1473-3099(16)00056-6
9. Shin J, Baek Y, Cho Y, et al. blaNDM-5-bearing IncFII-type plasmids of Klebsiella pneumoniae sequence type 147 transmitted by cross-border transfer of a patient. Antimicrob Agents Chemother. 2016;60(3):1932–1934. doi:10.1128/AAC.02722-15
10. Balm MND, La M, Krishnan P, Jureen R, Lin RTP, Teo JWP. Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin Microbiol Infect. 2013;19(9):E421–E423. doi:10.1111/1469-0691.12247
11. Bathoorn E, Rossen JW, Lokate M, Friedrich AW, Hammerum AM. Isolation of an NDM-5-producing ST16 Klebsiella pneumoniae from a Dutch patient without travel history abroad, August 2015. Euro Surveill. 2015;20:41. doi:10.2807/1560-7917.ES.2015.20.41.30040
12. Li A, Yang Y, Miao M, et al. Complete sequences of mcr-1-harboring plasmids from extended-spectrum-β-lactamase- and carbapenemase-producing enterobacteriaceae. Antimicrob Agents Chemother. 2016;60(7):4351–4354. doi:10.1128/AAC.00550-16
13. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing Twentieth Informational Supplement. Wayne, PA: CLSI; 2017.
14. The European Committee on Antimicrobial Susceptibility Testing. Break-Point Tables for Interpretation of MICs and Zone Diameters. European Committee on Antimicrobial Susceptibility Testing (EUCAST); 2017.
15. Endimiani A, Carias LL, Hujer AM, et al. Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae isolates possessing blaKPC in the United States. Antimicrob Agents Chemother. 2008;52(7):2680–2682. doi:10.1128/AAC.00158-08
16. Yang J, Chen Y, Jia X, et al. Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin Microbiol Infect. 2012;18(12):E506–E513. doi:10.1111/1469-0691.12035
17. Tsakris A, Pournaras S, Woodford N, et al. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J Clin Microbiol. 2000;38(3):1290–1292.
18. Poirel L, Héritier C, Tolün V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48(1):15–22. doi:10.1128/aac.48.1.15-22.2004
19. Senda K, Arakawa Y, Ichiyama S, et al. PCR detection of metallo-beta-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum beta-lactams. J Clin Microbiol. 1996;34(12):13.
20. Yu Y, Ji S, Chen Y, et al. Resistance of strains producing extended-spectrum beta-lactamases and genotype distribution in China. J Infect. 2007;54(1):53–57. doi:10.1016/j.jinf.2006.01.014
21. Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40(6):2153–2162. doi:10.1128/JCM.40.6.2153-2162.2002
22. Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178–4182. doi:10.1128/JCM.43.8.4178-4182.2005
23. Quan J, Li X, Chen Y, et al. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17(4):400–410. doi:10.1016/S1473-3099(16)30528-X
24. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–228. doi:10.1016/j.mimet.2005.03.018
25. Johnson TJ, Bielak EM, Fortini D, et al. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. 2012;68(1):43–50. doi:10.1016/j.plasmid.2012.03.001
26. Logan LK. Carbapenem-resistant enterobacteriaceae: an emerging problem in children. Clin Infect Dis. 2012;55(6):852–859. doi:10.1093/cid/cis543
27. Yu J, Tan K, Rong Z, et al. Nosocomial outbreak of KPC-2- and NDM-1-producing Klebsiella pneumoniae in a neonatal ward: a retrospective study. BMC Infect Dis. 2016;16(1):563. doi:10.1186/s12879-016-1987-z
28. Taoufik L, Hanchi AA, Fatiha B, et al. Emergence of OXA-48 carbapenemase producing Klebsiella pneumoniae in a neonatal intensive care unit in Marrakech, Morocco. Clin Med Insights Pediatr. 2019;13:1179556519834524. doi:10.1177/1179556519834524
29. Tian D, Pan F, Wang C, Sun Y, Zhang H. Resistance phenotype and clinical molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae among pediatric patients in Shanghai. Infect Drug Resist. 2018;11:1935–1943. doi:10.2147/IDR.S175584
30. Jian Z, Li Y, Liu W, et al. Detection of the novel IMP-38 among carbapenemase-producing Enterobacteriaceae in a university hospital, China. J Infect Dev Ctries. 2014;8(8):1044–1048. doi:10.3855/jidc.4179
31. Dong F, Lu J, Wang Y, et al. A five-year surveillance of carbapenemase-producing Klebsiella pneumoniae in a pediatric Hospital in China reveals increased predominance of NDM-1. Biomed Environ Sci. 2017;30(8):562–569. doi:10.3967/bes2017.075
32. Bi R, Kong Z, Qian H, et al. High prevalence of blaNDM variants among carbapenem-resistant Escherichia coli in Northern Jiangsu Province, China. Front Microbiol. 2018;9:2704. doi:10.3389/fmicb.2018.02704
33. Nour I, Eldegla HE, Nasef N, Shouman B, Abdel-Hady H, Shabaan AE. Risk factors and clinical outcomes for carbapenem-resistant Gram-negative late-onset sepsis in a neonatal intensive care unit. J Hosp Infect. 2017;97(1):52–58. doi:10.1016/j.jhin.2017.05.025
34. Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29(12):1099–1106. doi:10.1086/592412
35. Zhu J, Sun L, Ding B, et al. Outbreak of NDM-1-producing Klebsiella pneumoniae ST76 and ST37. Eur J Clin Microbiol Infect Dis. 2016;35(4):611–618. doi:10.1007/s10096-016-2578-z
36. Zhang W, Guo Y, Li J, et al. In vitro and in vivo bactericidal activity of ceftazidime-avibactam against Carbapenemase-producing Klebsiella pneumoniae. Antimicrob Resist Infect Control. 2018;7:142. doi:10.1186/s13756-018-0435-9
37. Zheng R, Zhang Q, Guo Y, et al. Outbreak of plasmid-mediated NDM-1-producing Klebsiella pneumoniae ST105 among neonatal patients in Yunnan, China. Ann Clin Microbiol Antimicrob. 2016;15:10. doi:10.1186/s12941-016-0124-6
38. Nakamura K, Kaneko M, Abe Y, et al. Outbreak of extended-spectrum beta-lactamase-producing Escherichia coli transmitted through breast milk sharing in a neonatal intensive care unit. J Hosp Infect. 2016;92(1):42–46. doi:10.1016/j.jhin.2015.05.002
39. Wailan AM, Paterson DL, Caffery M, Sowden D, Sidjabat HE. Draft genome sequence of NDM-5-producing Escherichia coli sequence type 648 and genetic context of blaNDM-5 in Australia Alexander. Genome Announc. 2015;3(2):e00194–15. doi:10.1128/genomeA.00194-15
40. Hammerum AM, Hansen F, Olesen B, et al. Investigation of a possible outbreak of NDM-5-producing ST16 Klebsiella pneumoniae among patients in Denmark with no history of recent travel using whole-genome sequencing. J Glob Antimicrob Resist. 2015;3(3):219–221. doi:10.1016/j.jgar.2015.07.001
41. Baraniak A, Izdebski R, Fiett J, et al. NDM-producing Enterobacteriaceae in Poland, 2012–14: inter-regional outbreak of Klebsiella pneumoniae ST11 and sporadic cases. J Antimicrob Chemother. 2016;71(1):85–91. doi:10.1093/jac/dkv282
42. Pitart C, Solé M, Roca I, et al. Molecular characterization of blaNDM-5 Carried on an IncFII plasmid in an Escherichia coli isolate from a nontraveler patient in Spain. Antimicrob Agents Chemother. 2015;59(1):659–662. doi:10.1128/AAC.04040-14
43. Nakano R, Nakano A, Hikosaka K, et al. First report of metallo-β-lactamase NDM-5-producing Escherichia coli in Japan. Antimicrob Agents Chemother. 2014;58(12):7611–7612. doi:10.1128/AAC.04265-14
44. Chen D, Gong L, Walsh TR, et al. Infection by and dissemination of NDM-5-producing Escherichia coli in China. J Antimicrob Chemother. 2016;71(2):563–565. doi:10.1093/jac/dkv352
45. Zhu YQ, Zhao JY, Xu C, Zhao H, Jia N, Li Y-N. Identification of an NDM-5-producing Escherichia coli sequence type 167 in a neonatal patient in China. Sci Rep. 2016;6:29934. doi:10.1038/srep29934
46. Kong L, Lei C, Ma S, et al. Various sequence types of Escherichia coli isolates coharboring blaNDM-5 and mcr-1 genes from a commercial swine farm in China. Antimicrob Agents Chemother. 2017;61(3):e02167–16. doi:10.1128/AAC.02167-16
47. He T, Wang Y, Sun L, Pang M, Zhang L, Wang R. Occurrence and characterization of blaNDM-5-positive Klebsiella pneumoniae isolates from dairy cows in Jiangsu, China. J Antimicrob Chemother. 2017;72(1):90–94. doi:10.1093/jac/dkw357
48. Liu B, Zhang X, Wan S, Hao -J-J, Jiang R-D, Song F-J. Characteristics of carbapenem-resistant Enterobacteriaceae in ready-to-eat vegetables in China. Front Microbiol. 2018;9:1147. doi:10.3389/fmicb.2018.01147
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.