Back to Journals » Infection and Drug Resistance » Volume 14
First Report of Oxacillin Susceptible mecA-Positive Staphylococcus aureus in a Children’s Hospital in Kunming, China
Authors Ma M, Chu M, Tao L, Li J, Li X, Huang H, Qu K , Wang H, Li L, Du T
Received 29 April 2021
Accepted for publication 25 June 2021
Published 6 July 2021 Volume 2021:14 Pages 2597—2606
DOI https://doi.org/10.2147/IDR.S317670
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Mingbiao Ma,1– 3,* Minjun Chu,1,2,* Lvyan Tao,3– 5,* Jue Li,1,2 Xiaojuan Li,1,2 Hailin Huang,1,2 Kexuan Qu,6 Haiping Wang,1,2 Li Li,3– 5 Tingyi Du1– 3
1Department of Clinical Laboratory, Kunming Children’s Hospital, Kunming, Yunnan, People’s Republic of China; 2Department of Clinical Laboratory, Children’s Hospital Affiliated to Kunming Medical University, Kunming, Yunnan, People’s Republic of China; 3Yunnan Key Laboratory of Children’s Major Disease Research, Kunming Children’s Hospital, Kunming, Yunnan, People’s Republic of China; 4Yunnan Institute of Pediatrics, Kunming Children’s Hospital, Kunming, Yunnan, People’s Republic of China; 5Kunming Key Laboratory of Children Infection and Immunity, Kunming Children’s Hospital, Kunming, Yunnan, People’s Republic of China; 6Department of Blood Transfusion, Kunming Children’s Hospital, Kunming, Yunnan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Li Li
Yunnan Institute of Pediatrics, Kunming Children’s Hospital, Kunming, Yunnan, People’s Republic of China
Email [email protected]
Tingyi Du
Department of Clinical Laboratory, Kunming Children’s Hospital, Kunming, Yunnan, People’s Republic of China
Email [email protected]
Purpose: The present study investigated the prevalence characteristics of oxacillin susceptible mecA-positive Staphylococcus aureus (OS-MRSA) in a children’s hospital in Kunming from January 2019 to December 2020.
Methods: A total of 499 S. aureus strains were included in the study and tested for oxacillin susceptibility using the VITEK 2 Compact automated antimicrobial susceptibility test system. All oxacillin-susceptible strains were detected mecA and mecC by polymerase chain reaction (PCR). E-test was used to compare the minimum inhibitory concentration (MIC) values of methicillin-susceptible S. aureus (MSSA), methicillin-resistant S. aureus (MRSA), and OS-MRSA for oxacillin, cefoxitin, penicillin, vancomycin, erythromycin, and clindamycin. Molecular typing of OS-MRSA was performed by MLST and SCCmec typing. Toxin genes were detected by PCR.
Results: Forty-five OS-MRSA strains were detected, for an overall rate of 9.02% (45/499). The MICs of MSSA, OS-MRSA, and MRSA against oxacillin were concentrated at 0.38, 0.38, and 12 μg/mL, respectively; the cefoxitin MICs of MSSA and MRSA were concentrated at 2 and 32 μg/mL respectively; and MICs of OS-MRSA were concentrated at 2 and 8 μg/mL; penicillin, vancomycin and erythromycin MICs against MSSA, OS-MRSA, and MRSA showed same centralized points and were 32, 1, and 256 μg/mL, respectively; the MICs of clindamycin against MSSA were 0.5 μg/mL, while that against OS-MRSA and MRSA were concentrated at 256 μg/mL. Molecular typing of OS-MRSA was dominated by ST59-SCCmec IV. The carrier rates of hemolysin genes (hl-a, hl-d) and fibrinogen-binding clumping factor genes (clfA, clfB) were 100% in OS-MRSA, followed by 40% (18/45) for enterotoxin genes (sea, seb).
Conclusion: OS-MRSA has a high detection rate in children, and main molecular typing is ST59-SCCmecIV in Kunming. The identification ability of automated antibacterial drug sensitivity test detection systems for OS-MRSA is very limited. A combination of phenotypic analysis and molecular detection should be used to improve OS-MRSA identification.
Keywords: Staphylococcus aureus, MRSA, OS-MRSA, children, oxacillin
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the common pathogens causing nosocomial and community-acquired infections, often leading to severe infections such as endocarditis, osteomyelitis, and sepsis. The mechanism of MRSA resistance to methicillin and other β-lactam antibiotics is mainly due to the carriage of mecA on the mobile genetic element staphylococcal chromosomal cassette mec (SCCmec), which encodes the synthetic low-affinity penicillin-binding protein PBP2a. Recent studies have found that the mecC gene also mediates drug resistance development.1
MRSA drug resistance has become a global issue of the high concern. The correct selection of antibiotics to treat MRSA infections in clinical practice depends on accurate laboratory identification of MRSA strains. Positive detection of the mecA or mecC gene is the “gold standard,” but its application in clinical laboratories is limited due to the complex, time-consuming, and expensive testing. At present, clinical laboratories generally use phenotypic analyses to identify MRSA. According to the recommendations of the Clinical and Laboratory Standards Institute (CLSI), they mainly include the minimum inhibitory concentration (MIC) method of oxacillin and cefoxitin. That is, oxacillin MIC ≤2 μg/mL or cefoxitin MIC ≤4 μg/mL is determined as methicillin-susceptible S. aureus (MSSA); oxacillin MIC ≥4 μg/mL or cefoxitin MIC ≥8 μg/mL is determined as MRSA. There is also the disk diffusion method with 30 μg cefoxitin; that is, an inhibition zone diameter ≥22 or ≤21 mm is determined as MSSA or MRSA, respectively.2 However, phenotypic analyses do not allow for the interpretation for strains with MICs between susceptible and resistant strains. More importantly, increasing studies have found that some clinical strains carry the mecA gene, but phenotypic analysis shows they are oxacillin or cefoxitin susceptible, named of Oxacillin-susceptible mecA gene-positive S. aureus (OS-MRSA).3–5 Routine phenotypic testing in clinical laboratories often misinterprets such strains as MSSA. Once these bacteria are exposed to β-lactam antibiotics, they are prone to induce MRSA with high resistance, resulting in clinical treatment failure.6,7 Therefore, identifying differences between OS-MRSA phenotypes and genotypes is challenging in clinical laboratory testing, and there are also risks for clinical empirical treatment.
There is a growing body of research on OS-MRSA in various regions and countries. Kampf et al7 showed that the detection rate of OS-MRSA was 1.6% in hospital healthcare workers, with six OS-MRSA strains that became highly resistant under repeated induction of oxacillin and cefotaxime. Only three true MRSA strains were detected in the present study, suggesting that S. aureus carried by healthcare workers may be insidious. Failure to identify such strains could lead to resistance development during antibiotic treatment, and then transmission of these strains to patients is more dangerous than MRSA. Petinaki et al8 detected OS-MRSA in up to 11.1% of Greek inpatients, suggesting that such strains might have been spreading but traditional detection methods failed to identify them. Hososaka et al9 found that OS-MRSA was detected in 1.25% of cases in Japan, and SCCmec typing showed that the main type was type III with molecular characteristics similar to community acquired MRSA. The detection rates were 3%, 10.9%, 1.17%, 1.2%, and 1.64–3.78% in Virginia (US), Taiwan (China), Iran, Wessex region (UK), and China, respectively.5,10–14 OS-MRSA strains are also detected in other animal species. Pu et al15 found a high detection rate (17.31–62.5%) in cow mastitis in many areas of China, suggesting that OS-MRSA strains may be closely related to this condition. Mistry and colleagues also confirmed the high detection rate of OS-MRSA in cow mastitis (48.71%) in India.13 OS-MRSA has also been detected in cheese and meat foods, which can cause food poisoning.16–18 Therefore, OS-MRSA is widely prevalent throughout the world, but the detection rate and molecular prevalence characteristics have significant geographical distribution differences.
Despite the growing number of reports on OS-MRSA in recent years, the relevant studies mainly focused on the prevalence and distribution of OS-MRSA in various regions and countries with case reports and molecular mechanism studies. Notably, several studies have shown that up to 60% of OS-MRSA strains are isolated from children, suggesting that the carrier rate may be higher in this population.4,11,19 However, no further research has assessed the specific prevalence of OS-MRSA in children, including molecular types, toxin genes carriage and resistance to commonly used antibiotics. Because children have an immature immune system and are more susceptible to pathogen infection than adults, it is of great clinical significance and value to understand the prevalence characteristics of OS-MRSA strains in children. The present study is the first investigation of the prevalence characteristics of OS-MRSA in children, including Multilocus sequence typing (MLST) and SCCmec typing, toxin genes carriage, and MIC distribution characteristics against commonly used antimicrobial agents.
Materials and Methods
Strain Collection and Identification
Duplicate samples obtained from the same site from the same patient were excluded, and 499 S. aureus strains were isolated from children visiting Kunming Children’s Hospital from January 2019 to December 2020 (229 in 2019 and 270 in 2020). Clinical sample types included sputum, lavage fluid, pus, various secretions, venous blood, and pleural effusion. Each strain was identified by colony morphology observation, Gram staining microscopy, and VITEK 2 Compact Automated Antimicrobial Susceptibility Test System card GP (bioMérieux, Marcy-l’Étoile, France).
VITEK 2 Compact Automated Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed according to the manufacturer instructions for the VITEK 2 Compact automated antimicrobial susceptibility test. Bacterial suspensions with a McFarland turbidity of 0.5 were prepared. The instrument automatically added suspension to the antimicrobial susceptibility test card GP-67. After incubation for 18–24h, the Advanced Expert SystemTM system could automatically read and analyze the susceptibility of various antibacterial drugs. The GP-67 card enabled cefoxitin screening and MIC test for oxacillin. S. aureus ATCC29213 was used as the quality control strain.
Polymerase Chain Reaction (PCR) Detection of mecA and mecC Genes in Oxacillin-Susceptible Strains
Based on the VITEK 2 Compact antimicrobial susceptibility test results, all oxacillin-susceptible strains were subjected to PCR for mecA and mecC gene detection. Bacterial genomic DNA was extracted using a bacterial genomic DNA extraction kit (TaKaRa Biotechnology, Shiga, Japan). The primer sequences of the mecA and mecC are listed in Table 1. PCR conditions were 94 °C for 15 min, 30 × (94 °C for 30 s, 59 °C for 1min, 72 °C for 1 min), and 72 °C for 10 min.20
 |
Table 1 The Main Primers Used for PCR Amplification in the Study |
E-Test to Compare the MICs of MSSA, OS-MRSA, and MRSA Strains
We randomly selected 30 MRSA strains and 30 MSSA strains for testing; all OS-MRSA strains were tested. We detected the MIC values against cefoxitin, oxacillin, penicillin, vancomycin, clindamycin, and erythromycin using E-test antimicrobial strips and Mueller-Hinton agar plates (Autobio Diagnostics, Zhengzhou, China). S. aureus ATCC29213 was used as a quality control strain, and the MIC50 and MIC90 represented the antibacterial drug concentrations that could inhibit the growth of 50% and 90% of bacteria, respectively.
MLST and SCCmec Molecular Typing of OS-MRSA Strains
All OS-MRSA strains were subjected to molecular typing by Multilocus sequence typing (MLST) and SCCmec typing. Seven housekeeping genes of MLST (aecC, aroE, glpF, gmk, pta, tpi, and yqiL) primer sequences and reaction conditions amplified by PCR were obtained from the MLST public database (https://pubmlst.org/). SCCmec typing performed as described previously.21
Toxin Genes Detection of OS-MRSA Strains
Toxin genes of OS-MRSA strains were detected by PCR, including hemolysin (hl-a, hl-d), fibrinogen-binding clumping factor (clfA, clfB), enterotoxin (sea, seb), Panton–Valentine leukocidin (pvl), and toxic shock syndrome 1 (tst-1); the primer sequences of toxin genes are shown in Table 1.22 The PCR amplification reaction conditions of pvl, tst-1, sea, and seb genes were 94 °C for 5 min, 30 × (94 °C for 30 s, 54 °C for 35 s, 72 °C for 1 min), and 72 °C for 10 min. Those for hl-a, hl-d, clfA, and clfB genes were: 94 °C for 5 min, 32 × (94 °C for 30 s, 57 °C for 35 s, 72 °C for 50 s), and 72 °C for 6 min.
Results
Sample Types and Patient Characteristics
The distribution of clinical sample types of 499 S. aureus strains were 239 (47.90%) from sputum; 114 (22.85%) from secretion; 90 (18.04%) from pus, and the remaining 56 (11.2%) from blood, bone marrow, Bronchoalveolar Lavage Fluid (BLAF) and pleural effusion. The patient group comprised 305 males and 194 females, and the mean age was 3.39 ± 4.28 years (range: 1 day to 17 years).
Identification of OS-MRSA Strains
The VITEK 2 compact detection results showed that among 499 S. aureus strains, 345 (166 strains in 2019, 179 strains in 2020) were susceptible to oxacillin, 45 (20 strains in 2019, 25 strains in 2020) were positive for the mecA gene by PCR. No strains were positive for mecC genes. Carriers of OS-MRSA strains were mainly children ≤1 year old, accounting for up 55.56% (25/45). The main types of clinical specimens were sputum accounting for 33.33% (15/45), secretions accounting for 31.11% (14/45), pus accounting for 24.44% (11/45), and a few isolated from blood and BLAF, accounting for 8.89% (4/45) and 2.22% (1/45), respectively. Most patients were from orthopedic surgery department accounting for 22.22% (10/45), followed by the intensive care unit (ICU) and burn (wound) plastic surgery department, both 17.78% (8/45), and neonatology 15.56% (7/45). The remaining strains were detected in other departments such as general surgery, respiratory medicine, and hematological oncology (Figure 1).
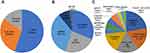 |
Figure 1 The distribution of patient ages, sample types and clinic departments among 45 OS-MRSA strains. (A) ages; (B) sample types; (C) clinic departments. |
MIC Distribution Characteristics of MSSA, OS-MRSA, and MRSA Strains
The MIC values of 30 randomly selected MSSA strains, 30 MRSA strains, and 45 OS-MRSA strains against oxacillin, cefoxitin, penicillin, vancomycin, clindamycin, and erythromycin were assessed with E-tests. The MICs of the experimental quality control strain S. aureus ATCC29213 against six antimicrobial agents were within the quality control ranges. The distributions of MIC values of MSSA, OS-MRSA, and MRSA strains against oxacillin mainly concentrated at 0.38, 0.38, and 12 μg/mL, respectively; the distributions of cefoxitin MICs of MSSA and MRSA were concentrated at 2 and 32 μg/mL respectively, and MICs of OS-MRSA were concentrated at 2 and 8 μg/mL; the distributions of penicillin, vancomycin, and erythromycin MICs against MSSA, OS-MRSA, and MRSA strains were mainly concentrated at 32, 1, and 256 μg/mL, respectively; the MIC of clindamycin against MSSA was 0.5 μg/mL, while those against OS-MRSA and MRSA were mainly concentrated at 256 μg/mL (Figure 2). Further analyses of the MIC50 and MIC90 results showed that the values against oxacillin and cefoxitin showed an upward trend for MSSA, OS-MRSA, and MRSA strains, as did the MIC50 values against penicillin and clindamycin (Table 2). No vancomycin-resistant strains were identified.
 |
Table 2 MIC50 and MIC90 Values of Three Kinds of S. aureus Strains Against Oxacillin, Cefoxitin, Penicillin, Vancomycin, Clindamycin, and Erythromycin |
 |
Figure 2 The MICs distribution of three kinds of S. aureus strains against oxacillin, cefoxitin, penicillin, vancomycin, clindamycin, and erythromycin. |
MLST and SCCmec Molecular Typing of OS-MRSA Strains
Except for three OS-MRSA strains for which the MLST database could not determine the ST type, the remaining 42 OS-MRSA strains were distributed in 14 different ST types, of which ST59 (11/45) was the most predominant ST type, followed by ST338 (8/45) and ST22 (5/45). SCCmec IV (44/45) was the most common SCCmec classification (Table 3). Therefore, ST59-SCCmec IV is the most common type of OS-MRSA strain from children in Kunming, followed by ST338-SCCmec IV.
 |
Table 3 Characteristics of 45 OS-MRSA Strains in the Study |
Toxin Genes Detection of OS-MRSA Strains
The results of PCR for the detection of five common toxin genes of S. aureus showed that the hemolysin (hl-a, hl-d) and fibrinogen-binding clumping factor (clfA and clfB) genes were present in 100% (45/45) of the OS-MRSA strains, followed by enterotoxin gene in 40% (18/45), which was mainly seb (17/18). The carrier rates of toxic shock syndrome (tsst-1) and Panton–Valentine leukocidin (pvl) genes were low at 6.67% (3/45) and 4.44% (2/45), respectively (Table 3)
Discussion
The present investigation was designed to assess the prevalence characteristics of OS-MRSA in children. To our knowledge, this is the first study on this topic. Previous reports on OS-MRSA were mainly in adult patients, even though some studies have shown that up to 60% of isolated OS-MRSA strains were from children, suggesting that that may have a higher carrier rate for this pathogen class. In the present study, the overall detection rate of OS-MRSA in children in Kunming was 9.02% for 2 years. The detection rate was 8.73% in 2019 and 9.30% in 2020 respectively, showing an increasing trend. The most common molecular type was ST59-SCCmecIV. Several groups have investigated the prevalence of OS-MRSA in China. He et al14 reported a detection rate of 3.78% that increased year by year (1.9 −4.8%). The molecular types of the strains were mainly ST59/338-t437-SCCmecIV/V. Song et al3 showed that the detection rate of OS-MRSA was 1.64% with the same molecular types of strains (mainly ST338/59-t437-SCCmecIV/V). The detection rate was slightly lower than that reported by He and colleagues, which may be due to the different cities included in the two studies. In addition, Liu et al19 showed that the detection rate of OS-MRSA was 1.8% in Shanghai, China, and the most common molecular type was ST59-SCCmecIV. Notably, 11 (64.71%) of the 17 OS-MRSA strains in that study were isolated from children. The results of Conceicao et al also showed that 60% of OS-MRSA strains were isolated from children.4 The molecular types detected in the present study were same with those reported in the relevant studies published in China, but the detection rate of OS-MRSA was significantly higher than that in the existing study reports. Possible reasons are as follows. (i) Previous relevant reports covered patients of all ages. These investigations focally reflected the overall prevalence of OS-MRSA in the region. Since the carrier rate of OS-MRSA may be different by age group, it could not accurately reflect the prevalence rates of such strains in children. (ii) The relevant studies showed that S. aureus ST59 was the main epidemic type in Asia, and the prevalence of ST59 was higher in children than in adults. This could lead to a higher carrier rate of OS-MRSA in children to a certain extent.23–25 (iii) Relevant studies reported that the antimicrobial susceptibility tests mainly used the broth microdilution method, which reveals higher susceptibility to MRSA.11,26 However, due to the complexity of the experiments and time required, this method is rarely used clinical laboratories, which typically use automated antimicrobial susceptibility test detection systems. MSSA screening in the present study was based on the VITEK 2 Compact, an automated antimicrobial susceptibility test system widely used in clinical laboratories, so the OS-MRSA detection rate is more in line with the actual clinical situation. In addition, the CLSI recommended the use of cefoxitin screening instead of oxacillin for MRSA identification in 2008. However, we found that 23 of 45 OS-MRSA strains were still negative for cefoxitin screening, so the identification ability of the automated antibacterial drug sensitivity test detection system for OS-MRSA was very limited. A combination of phenotypic analysis and molecular detection is required for accurate OS-MRSA identification.
In the present study, most OS-MRSA strains were isolated from children aged ≤1 year, which may be related to their immature immune systems that make them more susceptibility to pathogens. The main types of clinical specimens were sputum (33.33%), secretion (31.11%), and pus (24.44%), which were consistent with the findings of Liu et al.19 Patients were commonly seen in the orthopedic surgery department, followed by the ICU and burn (wound) plastic surgery. Since most of the secretion and pus samples came from the orthopedic surgery, ICU, and burn (wound) plastic surgery departments, the types of specimen were consistent with the distribution of departments. Taking into account the analysis of disease characteristics of patients, many patients treated in these departments undergo invasive procedures such as surgery, ventilation, gastric tube placement, tracheal intubation or incision that can damage the skin and respiratory mucosal barriers, facilitating S. aureus infection.
There are few reports on the distribution characteristics of OS-MRSA MICs against antimicrobial agents. In the present study, we analyzed and compared the MIC distribution characteristics of MSSA, OS-MRSA, and MRSA against commonly used antimicrobials (oxacillin, cefoxitin, penicillin, vancomycin, erythromycin, and clindamycin). The MIC distribution characteristics of oxacillin in the three kinds of S. aureus strains showed that all MSSA strains were in the susceptible range (MIC ≤2 μg/mL). Except for three strains of MRSA with MIC of 3 μg/mL, the MICs of the other strains were in the resistant range (MIC ≥4 μg/mL). The MICs of 33 strains of OS-MRSA were in the susceptible range, and 10 strains were in the resistant range, with corresponding MIC values mainly concentrated in 0.38 and 4 μg/mL, respectively. As expected, the MSSA and MRSA MICs against cefoxitin were in the susceptible (MIC ≤4 μg/mL) and resistant (MIC ≥8 μg/mL) ranges, respectively. However, the MIC values of OS-MRSA showed that half were susceptible and half were resistant, and there were also two centralized points of MIC values at 2 and 8 μg/mL. Oxacillin and cefoxitin are the antibacterial drugs recommended by the CLSI for clinical identification of MRSA. However, we found that there were two centralized points in the MIC values of OS-MRSA against these two drugs: one located in the susceptible range and one located at the susceptible cut-off point. Several groups reported that strains located at the cut-off point were often misinterpreted as MSSA, which was confirmed in the present study.27,28 There was no significant difference in the MIC distribution characteristics of MSSA, OS-MRSA, and MRSA strains against penicillin, vancomycin, and erythromycin. With regard to clindamycin MICs, the values of MSSA strains were mainly distributed at 0.5 μg/mL, while those of OS-MRSA were the same as those of MRSA, mainly concentrated at 256 μg/mL. Further analysis of the MIC50 and MIC90 of three types of strains against oxacillin, cefoxitin, penicillin, and clindamycin showed that the values of MSSA, OS-MRSA and MRSA strains against oxacillin and cefoxitin showed an upward trend, as did the MIC50 values against penicillin and clindamycin. This suggests that OS-MRSA may be a special type of S. aureus with a unique resistance profile, and the resistance range was between that of MSSA and MRSA. Controversy remains surrounding the treatment of OS-MRSA infection and whether β-lactam antibiotics can be used in clinical practice, and several studies have found that OS-MRSA strains can be induced to become highly resistant once exposed to β-lactam antibiotics, resulting in clinical treatment failure.6,7
Previous studies have shown that virulence factors play an important role in severe S. aureus infection, and the types and numbers of S. aureus strains carrying toxin genes are closely related to their invasiveness and pathogenicity.29,30 At present, there are very limited reports on the study of OS-MRSA virulence factors. We detected five common toxin genes of S. aureus and found that the carrier rates of the hemolysin (hl-a, hl-d) and fibrinogen-binding clumping factor (clfA, clfB) genes in OS-MRSA strains were up to 100% (45/45). The hemolysin gene is one of the most common toxins of S. aureus, and the hla and hld genes are important toxins causing S. aureus pneumonia and sepsis in clinical practice as they encode synthetic toxins that can destroy a variety of host cells and promote the development of inflammation. Fibrinogen-binding clumping factors encoded by the clfA and clfB genes are important clumping factors of S. aureus that can mediate binding to fibrin or fibrinogen of host cells to inhibit phagocytosis and promote bacterial invasion into the host, especially in the initial stage of infection. The Hososaka study also showed a high hemolysin gene carrier rate in OS-MRSA strains, but the distribution of fibrinogen-binding clumping factor genes in OS-MRSA strains has not been reported.9 In addition, we found that enterotoxin genes associated with food poisoning (sea, seb) had a high detection rate (40%) in OS-MRSA strains, mainly carrying seb. However, the six OS-MRSA strains studied by Hososaka et al9 were not found to carry enterotoxin genes. One study reported that 47% of MRSA strains in Chinese pediatric ICUs carried enterotoxin genes, similar to the results of the present study,31 while another group reported S. aureus in 68.5% of acquired bloodstream infections in Chinese children’s hospitals.32 Enterotoxin gene distributions may vary in different countries and regions with various population distributions and infection types. The low carrier rates of the toxic shock syndrome (tsst-1) and Panton–Valentine leukocidin gene (pvl) genes are consistent with the findings of Hososaka and colleagues.
Conclusion
We observed a high carrier rate (9.02%) of OS-MRSA in children, and the molecular typing was mainly ST59-SCCmecIV in Kunming. The MICs of OS-MRSA to antimicrobial agents are somewhat different from those of MSSA and MRSA and have unique distribution characteristics. Unfortunately, automated antibacterial drug sensitivity test detection system has very limited OS-MRSA identification ability. For children with S. aureus infection, it is important to determine the infected strain is OS-MRSA. Clinical laboratories should use a combination of phenotypic analysis and molecular detection to ensure accurate identification of these strains. This is the first study to describe the prevalence characteristics of OS-MRSA in children, but the limitations include the short surveillance duration and small sample size. Future prospective, longer-term investigations are needed to confirm the conclusion of the present study.
Ethical Approval
This study was conducted in accordance with the Declaration of Helsinki. The research protocol was approved by the Ethics Committee of Kunming Children’s Hospital (reference number 2020-03-020-K01). The Ethics Committee of Kunming Children’s Hospital did not require written informed consent from patients because the data of the patients used in the study were anonymized or maintained with confidentiality.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81960294, 82060291), the Basic Applied Study Planning Projects of Yunnan Province (No. 2021FB231), the Scientific Research Project of Education Department of Yunnan Province (No. 2020J0224), the Health Science and Technology Personnel Training Foundation of Kunming City of China (No. 2020-SW (reserve personnel)-109; 2021-SW (reserve personnel)-66; 2021-SW (reserve personnel)-72), the Science and Technology Planning Project of Yunnan Province (No. 2019ZF004-1-5-3).
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. García-álvarez L, Holden MTG, Lindsay H, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11(8):595–603. doi:10.1016/s1473-3099(11)70126-8
2. CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Eighth Informational Supplement. CLSI Document M100-S28. Wayne, PA: Clinical and Laboratory Standards Institute; 2018.
3. Song Y, Cui L, Lv Y, Li Y, Xue F. Characterisation of clinical isolates of oxacillin-susceptible mecA-positive Staphylococcus aureus in China from 2009 to 2014. J Glob Antimicrob Resist. 2017;11:1–3. doi:10.1016/j.jgar.2017.05.009
4. Conceicao T, Coelho C, de Lencastre H, Aires-de-sousa M. Frequent occurrence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) strains in two African countries. J Antimicrob Chemother. 2015;70(12):3200–3204. doi:10.1093/jac/dkv261
5. Saeed K, Ahmad N, Dryden M, et al. Oxacillin-susceptible methicillin-resistant Staphylococcus aureus (OS-MRSA), a hidden resistant mechanism among clinically significant isolates in the Wessex region/UK. Infection. 2014;42(5):843–847. doi:10.1007/s15010-014-0641-1
6. Kong LY, Jean A, Wong H, et al. Bacteremia caused by a mecA-positive oxacillin-susceptible Staphylococcus aureus strain with inducible resistance. Diagn Microbiol Infect Dis. 2015;83(4):377–378. doi:10.1016/j.diagmicrobio.2015.09.001
7. Kampf G, Adena S, Rüden H, Weist K. Inducibility and potential role of MecA-gene-positive oxacillin-susceptible Staphylococcus aureus from colonized healthcare workers as a source for nosocomial infections. J Hosp Infect. 2003;54(2):124–129. doi:10.1016/s0195-6701(03)00119-1
8. Petinaki E, Kontos F, Maniatis AN. Emergence of two oxacillin-susceptible mecA-positive Staphylococcus aureus clones in a Greek hospital. J Antimicrob Chemother. 2002;50(6):1090–1091. doi:10.1093/jac/dkf235
9. Hososaka Y, Hanaki H, Endo H, et al. Characterization of oxacillin-susceptible mecA-positive Staphylococcus aureus: a new type of MRSA. J Infect Chemother. 2007;13(2):79–86. doi:10.1007/s10156-006-0502-7
10. Forbes BA, Bombicino K, Plata K, et al. Unusual form of oxacillin resistance in methicillin-resistant Staphylococcus aureus clinical strains. Diagn Microbiol Infect Dis. 2008;61(4):387–395. doi:10.1016/j.diagmicrobio.2008.04.003
11. Chen FJ, Huang IW, Wang CH, et al. mecA-positive Staphylococcus aureus with low-level oxacillin MIC in Taiwan. J Clin Microbiol. 2012;50(5):1679–1683. doi:10.1128/JCM.06711-11
12. Jannati E, Arzanlou M, Habibzadeh S, et al. Nasal colonization of mecA-positive, oxacillin-susceptible, methicillin-resistant Staphylococcus aureus isolates among nursing staff in an Iranian teaching hospital. Am J Infect Control. 2013;41(11):1122–1124. doi:10.1016/j.ajic.2013.02.012
13. Mistry H, Sharma P, Mahato S, Saravanan R, Kumar PA, Bhandari V. Prevalence and characterization of oxacillin susceptible mecA-Positive clinical isolates of Staphylococcus aureus causing bovine Mastitis in India. PLoS One. 2016;11(9):e0162256. doi:10.1371/journal.pone.0162256
14. He W, Chen H, Zhao C, Zhang F, Wang H. Prevalence and molecular typing of oxacillin-susceptible mecA-positive Staphylococcus aureus from multiple hospitals in China. Diagn Microbiol Infect Dis. 2013;77(3):267–269. doi:10.1016/j.diagmicrobio.2013.07.001
15. Pu W, Su Y, Li J, et al. High incidence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) associated with bovine mastitis in China. PLoS One. 2014;9(2):e88134. doi:10.1371/journal.pone.0088134
16. Ariza-Miguel J, Oniciuc EA, Sanz I, Fernandez-Natal I, Hernandez M, Rodriguez-Lazaro D. Evaluation of two commercially available chromogenic media for confirmation of methicillin-resistant Staphylococcus aureus from human, animal, and food samples. Int J Food Microbiol. 2015;209:26–28. doi:10.1016/j.ijfoodmicro.2015.05.004
17. Luo R, Zhao L, Du P, et al. Characterization of an oxacillin-susceptible mecA-positive Staphylococcus aureus isolate from an imported meat product. Microb Drug Resist. 2020;26(2):89–93. doi:10.1089/mdr.2018.0211
18. Zhang P, Miao X, Zhou L, et al. Characterization of oxacillin-susceptible mecA-positive Staphylococcus aureus from food poisoning outbreaks and retail foods in China. Foodborne Pathog Dis. 2020;17(11):728–734. doi:10.1089/fpd.2019.2774
19. Liu JL, Li TM, Zhong N, et al. Current status of oxacillin-susceptible mecA-positive Staphylococcus aureus infection in Shanghai, China: a multicenter study. J Microbiol Immunol Infect. 2020. doi:10.1016/j.jmii.2020.07.021
20. Stegger M, Andersen PS, Kearns A, et al. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA(LGA251). Clin Microbiol Infect. 2012;18(4):395–400. doi:10.1111/j.1469-0691.2011.03715.x
21. Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43(10):5026–5033. doi:10.1128/JCM.43.10.5026-5033.2005
22. Qu WZ Study on drug resistance and virulence factors of Staphylococcu aureus isolated from clinical infected patients. 2019.Tianjin. Tianjin Medical University.
23. Geng W, Yang Y, Wu D, et al. Molecular characteristics of community-acquired, methicillinresistantStaphylococcus aureusisolated from Chinese children. FEMS Immunol Med Microbiol. 2010;58(3):356–362. doi:10.1111/j.1574-695X.2009.00648.x
24. Chen CJ, Unger C, Hoffmann W, Lindsay JA, Huang YC, Gotz F. Characterization and comparison of 2 distinct epidemic community-associated methicillin-resistant Staphylococcus aureus clones of ST59 lineage. PLoS One. 2013;8(9):e63210. doi:10.1371/journal.pone.0063210
25. Qiao Y, Ning X, Chen Q, et al. Clinical and molecular characteristics of invasive community-acquired Staphylococcus aureus infections in Chinese children. BMC Infect Dis. 2014;14:582. doi:10.1186/s12879-014-0582-4
26. Nair D, Shashindran N, Kumar A, Vinodh V, Biswas L, Biswas R. Comparison of phenotypic MRSA detection methods with PCR for mecA gene in the background of emergence of oxacillin-susceptible MRSA. Microb Drug Resist. 2021. doi:10.1089/mdr.2020.0361
27. Witte WWBPC, Pasemann B, Cuny C. Detection of low-level oxacillin resistance in mecA-positive Staphylococcus aureus.pdf. Clin Microbiol Infect. 2007;13:408–412. doi:10.1111/j.1469-0691.2006.01671.x
28. Felten A, Grandry B, Lagrange PH, Casin I. Evaluation of three techniques for detection of low-level methicillin-resistant Staphylococcus aureus (MRSA): a disk diffusion method with cefoxitin and moxalactam, the Vitek 2 system, and the MRSA-screen latex agglutination test. J Clin Microbiol. 2002;40(8):2766–2771. doi:10.1128/jcm.40.8.2766-2771.2002
29. Jenkins A, Diep BA, Mai TT, et al. Differential expression and roles of Staphylococcus aureus virulence determinants during colonization and disease. mBio. 2015;6(1):e02272–14. doi:10.1128/mBio.02272-14
30. Loughman JA, Fritz SA, Storch GA, et al. Virulence gene expression in human community-acquired Staphylococcus aureus infection. J Infect Dis. 2009;199(3):294–301. doi:10.1086/595982
31. Song Q, Wu J, Ruan P. Predominance of community-associated sequence type 59 methicillin-resistant Staphylococcus aureus in a paediatric intensive care unit. J Med Microbiol. 2018;67(3):408–414. doi:10.1099/jmm.0.000693
32. Yu F, Li T, Huang X, et al. Virulence gene profiling and molecular characterization of hospital-acquired Staphylococcus aureus isolates associated with bloodstream infection. Diagn Microbiol Infect Dis. 2012;74(4):363–368. doi:10.1016/j.diagmicrobio.2012.08.015
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
