Back to Journals » Infection and Drug Resistance » Volume 14
First Report of blaOXA-677 with Enhanced Meropenem-Hydrolyzing Ability in Pseudomonas aeruginosa in China
Authors Sun Y, Han R, Ding L, Yang Y, Guo Y, Wu S, Hu F, Yin D
Received 30 September 2021
Accepted for publication 9 December 2021
Published 31 December 2021 Volume 2021:14 Pages 5725—5733
DOI https://doi.org/10.2147/IDR.S340662
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Professor Suresh Antony
Yue Sun,1,2 Renru Han,1,2 Li Ding,1,2 Yang Yang,1,2 Yan Guo,1,2 Shi Wu,1,2 Fupin Hu,1,2 Dandan Yin1,2
1Institute of Antibiotics, Huashan Hospital, Fudan University, Shanghai, People’s Republic of China; 2Key Laboratory of Clinical Pharmacology of Antibiotics, Ministry of Health, Shanghai, People’s Republic of China
Correspondence: Fupin Hu; Dandan Yin Tel/Fax +86 21 52888186
; +86 21 52888196
Email [email protected]; [email protected]
Purpose: OXA-10-type class D β-lactamases have shown their evolutionary potential of enhancing carbapenem resistance. This study aimed to elucidate the role of OXA-10 variants in clinical isolated multidrug resistant (MDR) Pseudomonas aeruginosa and characterize the first appearance of OXA-677 in China.
Methods: Six blaOXA-10-like-positive strains were screened by PCR from 41 P. aeruginosa strains, which were resistant to both carbapenems and ceftazidime-avibactam, collected across China in 2018. The minimum inhibitory concentrations (MIC) were determined with the broth microdilution method. The resistance-associated genes and genetic environment were investigated by whole-genome sequencing (WGS). The function and mechanism of OXA-677 β-lactamase were identified by molecular cloning and protein structure modeling.
Results: All the blaOXA-10-like-positive Pseudomonas aeruginosa were MDR strains. They also had outer membrane porin defects and produced β-lactam resistance gene blaPER-1, fluoroquinolone-resistant gene crpP, aminoglycoside-resistance gene aph(3ʹ)-IIb, aph(6)-Id, aacA and aadA, fosfomycin-resistance gene fosA, sulfamethoxazole-resistance gene sul1, and chloramphenicol-resistance gene catB7. All blaOXA-10 variants were located in a Tn1403-related transposon, containing aacA4-12-blaOXA-677-aadA1, aacA4-12-blaOXA-101-aadA5, and blaOXA-246-aacA3-aadA13 gene cassette arrays, respectively. Notably, the blaOXA-677 producer showed a high MIC level of meropenem (MIC> 64 mg/L). Compared to blaOXA-10, blaOXA-677 was found a G-to-T transversion at position 350, leading to a phenylalanine-for-valine substitution in position 117, which is closer to leucine155 in the omega loop of the active site. MIC of meropenem for E. coli DH5α with the recombinant plasmid pHSG398 carrying blaOXA-677 was elevated by 8 times.
Conclusion: We speculate that the OXA-10-like enzymes and the decrease of membrane permeability confer carbapenem resistance, and the V117 substitution in OXA-677 might lead to a higher resistance level of meropenem.
Keywords: Pseudomonas aeruginosa, blaOXA-677, blaOXA-10-like gene, CHDL
Introduction
Pseudomonas aeruginosa is one of the most important and common clinical opportunistic pathogens, and its infection carries a high risk of mortality.1 Due to various inherent and acquired resistance mechanisms, the multidrug-resistant P. aeruginosa has emerged globally.2,3 There into, carbapenem-resistant P. aeruginosa was included in the highest category of urgency by the World Health Organization in 2017.4 According to the China Antimicrobial Surveillance Network (CHINET) of 2020, the resistance rates of P. aeruginosa to imipenem and meropenem are 23.2% and 19.3%, respectively.5
One or more mechanisms, including alteration of outer membranes, efflux pump overexpression, and production of carbapenemases, have been found contributing to the carbapenem resistance in P. aeruginosa isolates.6–8 It is generally considered that only enzymes with broad-spectrum and significant carbapenem hydrolysis activities are the most predominant carbapenemases, such as the class B metal-β-lactamases NDM, IMP, VIM, and the class A serine-β-lactamases KPC. In recent years, special attention has been given to the class D serine-β-lactamase OXA-family.
The OXA-family counts 926 members and only several groups are considered as carbapenem-hydrolyzing class D β-lactamases (CHDLs), such as OXA-23-like, OXA-40/24-like, OXA-58-like, and OXA-48-like β-lactamases. Among these carbapenemases, only OXA-40-like genes have been identified in P. aeruginosa, and the rest are prevalent in A. baumannii. However, some studies suggest that OXA-10 and OXA-2 β-lactamases can also confer reduced susceptibility to carbapenems when they express in strains with outer membrane porin defects, just like the well-known CHDL OXA-48.9 Furthermore, modification of OXA-10 may enhance resistance to either carbapenem or new β-lactam/β-lactamase inhibitor combination.10,11 According to the NCBI database, a total of 37 OXA-10-like enzymes have been reported around the world so far, and over 75% (28/27) of them have been detected in P. aeruginosa.12–14 OXA-14, OXA-794, OXA-795, and OXA-824, confer resistance to β-lactam/β-lactamase inhibitor but decrease meropenem MICs.10 Kotsakis et al reported the first natural variant OXA-655 (V117L) with increased carbapenem hydrolysis produced by an environmental E. coli strain.15 In this study, a variant similar to OXA-655 (OXA-677, V117F) and other two variants (OXA-101 and OXA-246) were found to be produced by several clinically relevant carbapenem resistant P. aeruginosa isolates. Here, we tried to characterize the rare variant OXA-677 and elucidate the role of OXA-10 variants in carbapenem resistance.
Materials and Methods
Strains and Plasmid
P. aeruginosa strains named 18-W18-009, 18-W18-012, 18-W71-012, 18-W71-013, 18-W71-020, 18-W31-087 were found producing OXA-10-like enzymes during the screen for clinical isolates, which were resistant to both carbapenems and ceftazidime-avibactam among 39 strains by the CHINET workgroup in 2018. All strains were identified using VITEK matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS, bioMérieux, Marcyl’Étoile, France). P. aeruginosa ATCC 27853 was used as the quality control strain for antimicrobial susceptibility testing; P. aeruginosa PAO1 was used as the reference strain for the whole-genome sequence blasting in the Genebank; E. coli DH5α and plasmid pHSG398 were used as the recipient bacterium for transformation test and the resistance gene carrier for clone experiment, respectively.
Antimicrobial Susceptibility Testing
Minimal inhibitory concentrations (MICs) were determined with broth microdilution method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines of the 30th edition. Antimicrobial agents used in antimicrobial susceptibility tests were imipenem, meropenem, ceftazidime/avibactam, piperacillin/tazobactam, ceftolozane/tazobactam, cefepime, amikacin, polymyxin B, ceftazidime, aztreonam, and ciprofloxacin. MIC interpretation for most agents was performed based on the CLSI breakpoints except for polymyxin, which was interpreted using EUCAST standards.16
Whole Genome Sequencing and Bioinformatics Analyses
Total genomic DNA was extracted using the genomic DNA extraction kit (Tiangen, China). Fragmented genomic DNA (~400-bp) was obtained through the Covaris S2 (Covaris, USA) and library construction was performed by NEXTflex DNA Sequencing Kit compatible with the Biomek® FXp (Bio Scientific, USA). Genome-wide sequencing was performed using Illumina Solexa technology (Illumina, San Diego, CA, USA). De novo assembly was carried out with Velvet v1.2.08 and redundant contigs were removed using the Fastx toolkit.
The multilocus sequence typing (MLST) was identified by an online Pseudomonas aeruginosa mlst typing database (https://pubmlst.org/). The presence of horizontally acquired resistance mechanisms was detected with the ResFinder v4.1 server (https://cge.cbs.dtu.dk//services/ResFinder/) and the whole bacterial genome annotation was performed by Rapid Annotation using Subsystem Technology (RAST) (https://rast.nmpdr.org/). The mobile element-related genes (MGEs) were identified by MobileElementFinder (https://cge.cbs.dtu.dk/services/MobileElementFinder/), ISfinder (https://www-is.biotoul.fr), and Tn Number Registry (https://transposon.lstmed.ac.uk/). Additionally, gene alterations and disruptions of porin-encoding genes oprD and efflux pump repressor gene mexR, nalC, nalD were compared with those of P. aeruginosa PAO1 using SnapGene.
Molecular Cloning, Transformation Test and Protein Structure Modeling of blaOXA-677
The gene of blaOXA-677 was amplified with Q5® High-Fidelity 2X Master Mix. OXA-10-F-EcoRI (5ʹ-CGGAATTCGCCATGAAAACATTTGCCGCATATG-3ʹ) and OXA-10-R-BamHI (5ʹ -CGCGGATCCGTTAGCCACCAATGATGCCC-3ʹ) were used as the primer pair. The cycling parameters were 98°C for 30 s, 30 cycles at 98°C for 30 s, 63°C for 30 s and 72°C for 60 s. The PCR product was ligated to the vector plasmid pHSG398 and then the recombinant plasmid was transferred into the recipient strain E. coli DH5α. Mueller-Hinton agar plates containing 50 mg/L of chloramphenicol (for pHSG398) and 50 mg/L of amoxicillin (for blaOXA-677) were used to select the potential transformants, which were confirmed finally by PCR and sanger sequencing. The three-dimensional protein structure models for OXA-677 carbapenemase were built by SWISS-MODEL Server (https://swissmodel.expasy.org/) and PyMOL software (https://www.schrodinger.com/products/pymol), using that of OXA-655 carbapenemase (PDB accession number 6SKQ; resolution, 2.10Å) as a reference.
Nucleotide Accession Numbers
The sequences of OXA-10 variants and their gene context have been deposited in the GenBank database under accession numbers OL353898 (blaOXA-101), OL353899 (blaOXA-246) and OL353897 (blaOXA-677).
Results
Resistance Phenotypes and Genotypes of blaOXA-10 Variants Positive Strains
P. aeruginosa strains named 18-W18-009, 18-W18-012, 18-W71-012, 18-W71-013, 18-W71-020, 18-W31-087 expressed multidrug resistant phenotypes. They were all resistant to imipenem, meropenem, ceftazidime/avibactam, piperacillin/tazobactam, ceftolozane/tazobactam, cefepime, amikacin, ceftazidime, aztreonam, and ciprofloxacin. P. aeruginosa named 18-W31-087 showed a higher MIC level of meropenem (MIC > 64 mg/L) than others (MIC = 8–32 mg/L). No carbapenemase gene could be detected except blaOXA-10-like in these strains by the PCR (all carbapenemase genes tested in this study were shown in Table S1 with primers sequence), as well as the following whole-genome sequencing (WGS). PCR and WGS data revealed that strain 18-W31-087 produced OXA-677 enzyme, strain 18-W18-009 and 18-W18-012 produced OXA-256 enzyme, strain 18-W71-012, 18-W71-013, 18-W71-020 produced OXA-101 enzyme. blaOXA-246-positive strains belonged to ST298, and others belonged to ST244. Meanwhile, chromosomal mutations such as premature termination of the efflux pump repressor gene mexR and porin-encoding gene oprD were observed in blaOXA-246-positive strains. And for strains producing OXA-677 or OXA-101 enzymes, premature stop codon occurred in oprD genes. Besides the carbapenem resistance associated genes, all the six strains produced extended spectrum β-lactamase (ESBL) gene blaPER-1, fluoroquinolone-resistant gene crpP, aminoglycoside-resistance gene aph(3ʹ)-IIb, aph(6)-Id, aacA and aadA, fosfomycin-resistance gene fosA, sulfamethoxazole-resistance gene sul1, and chloramphenicol-resistance gene catB7. blaOXA-246-positive strains also produced blaTEM-1B and aminoglycoside-resistance gene rmtB (Table 1).
 |
Table 1 Demographic, Antimicrobial Susceptibility and Genotypic Data for the Clinical Isolates of P. aeruginosa Harboring OXA-10 Natural Variants |
Genetic Context and Protein Structure of blaOXA-10 Variants
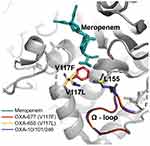
All three kinds of blaOXA-10 variants are located in a Tn1403-related transposon of the Tn-3 family. Each transposon contains a complex class 1 integron harboring an ISCR1-blaPER-1 unit. The difference of the three class 1 integrons was gene cassette arrays (GCA) in the variable region (VR). GCA aacA4-12-blaOXA-677-aadA1, aacA4-12-blaOXA-101-aadA5, and blaOXA-246-aacA3-aadA13 were detected, respectively (Figure 1). Compared to the blaOXA-10, the sequence of blaOXA-677 was found carrying a G-to-T transversion at position 350, leading to a phenylalanine-for-valine substitution in position 117 (Val117Phe); whereas there are no base mutations for either blaOXA-101 or blaOXA-246 at the key amino acid motifs (Figure 2). Val117Phe (V117F) was located at the second catalytically important conserved motif of class D β-lactamases. Due to the substitution, the phenylalanine117 is closer to leucine155 in the omega loop (Figure 3).
 |
Figure 1 The genomic context of the blaOXA-677 (A), blaOXA-101 (B), blaOXA-246 (C). Genes are denoted by arrows. Genes, MGEs, and other features are colored based on their functional classification. |
Characteristic of Transformants Carrying blaOXA-677
Transformants were obtained from the screening plates after verification of an 822bp PCR-amplified product of blaOXA-677. Two sequence-confirmed transformants were randomly selected for antimicrobial susceptibility tests to ensure consistent results. Compared with the empty host, MIC of meropenem for the E. coli DH5α transformant with the recombinant plasmid pHSG398 carrying blaOXA-677 was elevated by 8 times (Table 2). No significant increase was observed for imipenem, as well as ceftolozane/tazobactam, piperacillin/tazobactam, ceftazidime/avibactam, ceftazidime, aztreonam, cefepime, amikacin, polymyxin B, and ciprofloxacin (Table 2).
 |
Table 2 β-Lactam Resistance Profile Conferred by OXA-677 in E. coli DH5α Under Isogenic Conditions |
Discussion
OXA-10-like enzymes are widespread in P. aeruginosa and A.baumannii, and less common in Enterobacterales in Asia, Europe, South America, and Africa. It is the first report of the existence of blaOXA-10-like in ST298 and ST244 clones in China, which are two of the top 10 high-risk MDR/XDR P. aeruginosa clones causing nosocomial infection worldwide.17 blaOXA-101 was first reported in Enterobacteriaceae, Citrobacter freundii, Escherichia coli, and Enterobacter cloacae in Argentina in 2005 and now has been detected in A. baumannii and P. aeruginosa in China.18–20 blaOXA-246 was first found among ceftazidime-resistant P. aeruginosa clinical isolates in China in 2007.21 blaOXA-677 and its GCA aacA4-12-blaOXA-677-aadA1 were found for the first time in China in this study. Our study showed that these three variants were all embedded in class 1 integrons harboring an ISCR1-blaPER-1 unit. In public databases, the most similar integron was In1079, located on the chromosome of P. aeruginosa.22
Previous studies showed that the lack of OprD protein could only raise meropenem to 2–4 mg/L.23 Antunes et al suggested that the OXA-10 β-lactamase, which is regarded as the narrow-spectrum enzyme, is CHDL in fact.9 We speculate that the production of OXA-10-like enzymes and decrease of membrane permeability may cooperate kindly to confer meropenem resistance, for we found premature termination of oprD genes for all six blaOXA-10 variants positive strains in this study. Moreover, the antimicrobial susceptibility data of the clinical isolates and transformants showed that blaOXA-677 producing strain had a higher MIC level of meropenem. However, there was no difference between the resistance genes of blaOXA-677-positive strain and blaOXA-101-positive strains other than blaOXA-10-like genes. Structural bases of OXA-677 may explain the high-level resistance. As previously shown, OXA-677 has a single mutation at Val117. The activity of class D β-lactamases, including OXA-10, is dependent on Lys70 carboxylation in the active site. Val117 of the S115A116V117 catalytic motif is the component of the hydrophobic core surrounding Lys70 and is strictly conserve.24,25 It was validated in OXA-655 with V117L that alterations of the interactions with carbapenems were brought by substitutions at this key site.26 In both crystal structures of OXA-655 and OXA-677 proteins, their position 117 residues are closer to Leu155 in the omega loop, and their interactions seem stronger. This conformational change probably alters the accessibility of the meropenem to the active site. Further work is required to demonstrate the correlation between its structure and function.
In addition, all OXA-10 variants-producing isolates in this study harbor ISCR1-blaPER-1. Recent studies have shown that blaPER-1 may be a source of resistance to the new class of β-lactam/β-lactamase inhibitors ceftazidime/avibactam and ceftolozane/tazobactam.27 Therefore, these β-lactamase genes combined with porin loss may confer resistance to almost all kinds of carbapenem and β-lactam/β-lactamase inhibitors. Coupled with other gene cassettes (aadA, aacA, sul1) in Tn3-like transposons conferring resistance to fluoroquinolone, aminoglycosides, and sulfonamides, these clinical isolates may be more likely to develop to MDR/XDR strains. And these gene cassettes might be transmitted among different strains due to the mobile genetic element. It is unfortunate from a clinical treatment standpoint for there might be no antibiotic available for these strains. Polymyxin B might be the only treatment option, albeit with toxicity. Therefore, screening for OXA-10 mutants during the treatment of P. aeruginosa infection may have some instructive significance for adjustment for medication use.
Conclusion
In summary, globally widespread OXA-10-like enzymes have shown their evolutionary potential toward enhancing carbapenem resistance. In this study, OXA-677 with increased carbapenemase activity was identified in a clinical MDR P. aeruginosa strain for the first time in China. It is also the first report of the existence of OXA-10 variants (OXA-677, OXA-101, and OXA-246) in ST298 and ST244 clones in China. Our findings provide evidence supporting the claim that OXA-10 should be classified as CHDL. It may help elucidate the role of OXA-10 variants in MDR P. aeruginosa strains. blaOXA-10-like along with other gene cassettes in class 1 integrons and membrane permeability defects contribute to the resistance to almost all available antibiotics, which brings challenges in their treatment. Further work with a larger number of positive strains is now needed to elucidate the mechanism of OXA-677 and other OXA-10 mutants.
Ethics Approval
The study protocol was approved by the Institutional Review Board of Huashan Hospital, Fudan University (No. 2020-041).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81871690 and 81902101), and China Antimicrobial Surveillance Network (2020QD049).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Moradali MF, Ghods S, Rehm BH. Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front Cell Infect Microbiol. 2017;7:39. doi:10.3389/fcimb.2017.00039
2. Perez A, Gato E, Perez-Llarena J, et al. High incidence of MDR and XDR Pseudomonas aeruginosa isolates obtained from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J Antimicrob Chemother. 2019;74(5):1244–1252. doi:10.1093/jac/dkz030
3. Yin D, Wu S, Yang Y, et al. Results from the China Antimicrobial Surveillance Network (CHINET) in 2017 of the in vitro activities of ceftazidime-avibactam and ceftolozane-tazobactam against clinical isolates of Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2019;63(4):e02431–18. doi:10.1128/AAC.02431-18
4. Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi:10.1016/S1473-3099(17)30753-3
5. Hu F, Guo Y, Zhu D, et al. CHINET surveillance of bacterial resistance: results of 2020. Chin J Infect Chemother. 2021;21(04):377–387.
6. Shen J, Pan Y, Fang Y. Role of the outer membrane protein OprD2 in carbapenem-resistance mechanisms of Pseudomonas aeruginosa. PLoS One. 2015;10(10):e0139995. doi:10.1371/journal.pone.0139995
7. Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis. 2002;34(5):634–640. doi:10.1086/338782
8. López-Causapé C, Cabot G, Del Barrio-Tofiño E, Oliver A. The versatile mutational resistome of Pseudomonas aeruginosa. Front Microbiol. 2018;9:685. doi:10.3389/fmicb.2018.00685
9. Antunes NT, Lamoureaux TL, Toth M, et al. Class D β-lactamases: are they all carbapenemases? Antimicrob Agents Chemother. 2014;58(4):2119–2125. doi:10.1128/AAC.02522-13
10. Arca-Suárez J, Lasarte-Monterrubio C, Rodiño-Janeiro B, et al. Molecular mechanisms driving the in vivo development of OXA-10-mediated resistance to ceftolozane/tazobactam and ceftazidime/avibactam during treatment of XDR Pseudomonas aeruginosa infections. J Antimicrob Chemother. 2021;76(1):91–100. doi:10.1093/jac/dkaa396
11. De Luca F, Benvenuti M, Carboni F, et al. Evolution to carbapenem-hydrolyzing activity in noncarbapenemase class D β-lactamase OXA-10 by rational protein design. Proc Natl Acad Sci Am. 2011;108(45):18424–18429. doi:10.1073/pnas.1110530108
12. Gu B, Tong M, Zhao W, et al. Prevalence and characterization of class I integrons among pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J Clin Microbiol. 2007;45(1):241–243. doi:10.1128/JCM.01318-06
13. Farajnia S, Ansarin K, Mohsenchian A. Prevalence of OXA-2 and OXA-10 type ESBL and class I integron among Acinetobacter baumannii strains isolated from patients in Tabriz–North West Iran. Int J Infect Dis. 2012;16:e414. doi:10.1016/j.ijid.2012.05.567
14. Yoon EJ, Jeong SH. Class D beta-lactamases. J Antimicrob Chemother. 2021;76(4):836–864. doi:10.1093/jac/dkaa513
15. Kotsakis SD, Flach C, Razavi M, Larsson DGJ. Characterization of the first OXA-10 natural variant with increased carbapenemase activity. Antimicrob Agents Chemother. 2019;63(1). doi:10.1128/AAC.01817-18
16. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 11.0; 2021. Available from: http://www.eucast.org/clinical_breakpoints/.
17. Del Barrio-Tofino E, Lopez-Causape C, Oliver A, et al. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int J Antimicrob Agents. 2020;56(6):106196. doi:10.1016/j.ijantimicag.2020.106196
18. Porto A, Ayala J, Gutkind G, Di Conza J. A novel OXA-10–like β-lactamase is present in different Enterobacteriaceae. Diagn Microbiol Infect Dis. 2010;66(2):228–229. doi:10.1016/j.diagmicrobio.2009.09.010
19. Liu M, Ma J, Jia W, Li W. Antimicrobial resistance and molecular characterization of gene cassettes from class 1 Integrons inPseudomonas aeruginosa strains. Microb Drug Resist. 2020;26(6):670–676. doi:10.1089/mdr.2019.0406
20. Juan C, Zamorano L, Pérez JL, Ge Y, Oliver A. Activity of a new antipseudomonal cephalosporin, CXA-101 (FR264205), against carbapenem-resistant and multidrug-resistant Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother. 2010;54(2):846–851. doi:10.1128/AAC.00834-09
21. Qing Y, Cao K, Fang Z, et al. Outbreak of PER-1 and diversity of β-lactamases among ceftazidime-resistant Pseudomonas aeruginosa clinical isolates. J Med Microbiol. 2014;63(3):386–392. doi:10.1099/jmm.0.069427-0
22. Yu T, Yang H, Li J, et al. Novel chromosome-borne accessory genetic elements carrying multiple antibiotic resistance genes in Pseudomonas aeruginosa. Front Cell Infect Microbiol. 2021;11:638087. doi:10.3389/fcimb.2021.638087
23. Livermore DM. Of Pseudomonas, porins, pumps and carbapenems. J Antimicrob Chemother. 2001;47(3):247–250. doi:10.1093/jac/47.3.247
24. Vercheval L, Bauvois C, Di Paolo A, et al. Three factors that modulate the activity of class D β-lactamases and interfere with the post-translational carboxylation of Lys70. Biochem J. 2010;432(3):495–506. doi:10.1042/BJ20101122
25. Maveyraud L, Golemi D, Kotra LP, et al. Insights into class D beta-lactamases are revealed by the crystal structure of the OXA-10 enzyme from Pseudomonas aeruginosa. Structure. 2000;8(12):1289–1298. doi:10.1016/S0969-2126(00)00534-7
26. Leiros HKS, Thomassen AM, Samuelsen Ø, et al. Structural insights into the enhanced carbapenemase efficiency of OXA‐655 compared to OXA‐10. Febs Open Bio. 2020;10(9):1821–1832. doi:10.1002/2211-5463.12935
27. Ortiz De La Rosa J, Nordmann P, Poirel L. ESBLs and resistance to ceftazidime/avibactam and ceftolozane/tazobactam combinations in Escherichia coli and Pseudomonas aeruginosa. J Antimicrob Chemother. 2019;74(7):1934–1939. doi:10.1093/jac/dkz149
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.