Back to Journals » Journal of Inflammation Research » Volume 17
FBL Promotes LPS-Induced Neuroinflammation by Activating the NF-κB Signaling Pathway
Authors Zhang Z, Liu D, Lv R, Zhao H, Li T, Huang Y , Tian Z, Gao X, Luo P, Li X
Received 21 November 2023
Accepted for publication 2 April 2024
Published 11 April 2024 Volume 2024:17 Pages 2217—2231
DOI https://doi.org/10.2147/JIR.S451049
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tara Strutt
Zhuoyuan Zhang,1,2,* Dan Liu,1,2,* Rui Lv,1,2,* Haoyan Zhao,1,2,* Tianjing Li,1,2 Yutao Huang,2 Zhicheng Tian,2 Xiangyu Gao,2 Peng Luo,2 Xin Li3
1Biochemistry and Molecular Biology, College of Life Science, Northwest University, Xi’an, 710127, People’s Republic of China; 2Department of Neurosurgery, Xijing Hospital, Fourth Military Medical University, Xi’an, 710032, People’s Republic of China; 3Department of Anesthesiology, Xijing Hospital, Fourth Military Medical University, Xi’an, 710032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Peng Luo; Xin Li, Email [email protected]; [email protected]
Purpose: Neuroinflammation occurs in response to central nervous system (CNS) injury, infection, stimulation by toxins, or autoimmunity. We previously analyzed the downstream molecular changes in HT22 cells (mouse hippocampal neurons) upon lipopolysaccharide (LPS) stimulation. We detected elevated expression of Fibrillarin (FBL), a nucleolar methyltransferase, but the associated proinflammatory mechanism was not systematically elucidated. The aim of this study was to investigate the underlying mechanisms by which FBL affects neuroinflammation.
Methods: RT-real-time PCR, Western blotting and immunofluorescence were used to assess the mRNA and protein expression of FBL in HT22 cells stimulated with LPS, as well as the cellular localization and fluorescence intensity of FBL. BAY-293 (a son of sevenless homolog 1 (SOS1) inhibitor), SR11302 (an activator protein-1 (AP-1) inhibitor) and KRA-533 (a KRAS agonist) were used to determine the molecular mechanisms underlying the effect of FBL. AP-1 was predicted to be the target protein of FBL by molecular docking analysis, and validation was performed with T-5224 (an AP-1 inhibitor). In addition, the downstream signaling pathways of FBL were identified by transcriptome sequencing and verified by RT-real-time PCR.
Results: LPS induced FBL mRNA and protein expression in HT22 cells. In-depth mechanistic studies revealed that when we inhibited c-Fos, AP-1, and SOS1, FBL expression decreased, whereas FBL expression increased when KRAS agonists were used. In addition, the transcript levels of inflammatory genes in the NF-kB signaling pathway (including CD14, MYD88, TNF, TRADD, and NFKB1) were elevated after the overexpression of FBL.
Conclusion: LPS induced the expression of FBL in HT22 cells through the RAS/MAPK signaling pathway, and FBL further activated the NF-kB signaling pathway, which promoted the expression of relevant inflammatory genes and the release of cytokines. The present study reveals the mechanism by which FBL promotes neuroinflammation and offers a potential target for the treatment of neuroinflammation.
Keywords: FBL, neuroinflammation, LPS, molecular docking, transcriptome sequencing
Introduction
Inflammation is a response to damage or trauma to or infection of cells or tissues, and inflammation that occurs in the brain is called neuroinflammation. Neuroinflammation is an immune response characterized by overactivation of glia in the central nervous system (CNS). This leads to the activation of inflammatory cells in the brain, including microglia and astrocytes, and the release of cytokines and the production of free radicals can lead to neuronal cell death and synaptic dysfunction. Mild neuroinflammation is conducive to neuronal repair and maintenance, while continued uncontrolled glial activation and microenvironment destruction can exacerbate neuronal damage and death in aging and neurodegenerative diseases.1 Therefore, dysregulation of the inflammatory response may lead to brain damage.2
Neuroinflammation is an integral process in neurodegenerative diseases. Transient neuroinflammatory signaling plays a protective role during development and tissue repair after injury, whereas chronic neuroinflammation is associated with the progression of neurodegenerative diseases such as Alzheimer’s disease (AD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS) and multiple sclerosis (MS).3–5 The main commonalities between neuroinflammation that occurs in these diseases and inflammation induced by LPS are the significant activation of Toll-like receptor 4 (TLR4) signaling and the production of potent proinflammatory cytokines. TLR4 is the most well characterized member of the TLR family and the only receptor that uses all four INTER domain-containing connectors. Activation of TLR4 in response to LPS induces two signaling pathways, termed the myd88-dependent pathway and the myd88-independent pathway. The Myd88-dependent pathway requires MyD88 and TIRAP to activate NF-κB and proinflammatory cytokine production. Myd88-independent signaling events are controlled by TRIF and TRAM and induce IRF3-dependent type I interferon production.6 To model the complexity of processes involving neuroinflammation and their outcomes, such as cognitive dysfunction, in vivo studies are often used. LPS is one of the most potent proinflammatory stimuli.7,8 LPS-induced inflammation is widely used in a range of assays to study interference with inflammatory pathways.
It is well known that the transcription of inflammatory cytokines is regulated by several key transcription factors, including nuclear factor kB (NF-kB),9 activator protein-1 (AP-1),10 nuclear factors of activated T cell (NFATs),11 and signal transducer and activator of transcription (STAT).12 Various reports have shown that the activation of mitogen-activated protein kinases (MAPKs), including extracellular signal-regulated kinases (ERKs), c-Jun N-terminal kinases (JNK), and p38, can promote the expression and phosphorylation of c-Jun and c-Fos, which form AP-1 dimers during inflammatory responses and induce the transcription of proinflammatory proteins.13 However, NF-kB is considered the main transcription factor that regulates proinflammatory mediators.14 When HT22 cells are in the resting state, NF-kB is located in the cytoplasm as a dimer consisting of the p50 and p65 subunits. In response to LPS stimulation, NF-κB inhibitors phosphorylate and degrade NF-κB dimers, which are activated and translocate into the nucleus, leading to the transcription of proinflammatory factors such as NLR family pyrin structural domain containing 3 (NLRP3). Therefore, inhibiting NF-kB activation is widely recognized as an effective therapeutic strategy for reducing neuroinflammation.15
There is increasing evidence that the nucleolus plays a role in regulating cellular processes related to health and disease. FBL is a highly conserved nucleolus methyltransferase that is an important factor in the C/D small nucleolar ribonucleoprotein (snoRNP) complex. FBL mediates the methylation of the 2′-O-ribose of rRNAs, thereby contributing to rRNA maturation.16 FBL also methylates histone H2A, which acts as a protein methyltransferase.17 Many studies have shown that FBL plays an important role in regulating processes such as the cellular stress response, but its regulatory mechanism in the inflammatory response has not been elucidated.
In this study, we found that LPS significantly increased FBL protein expression in HT22 cells, leading to inflammation. Further evidence suggested that upstream FBL was activated by the RAS signaling pathway and subsequently regulated inflammatory responses via the NF-kB pathway. These results suggest that FBL plays an important mediating role in the proinflammatory immune response in HT22 cells, ultimately participating in the pathogenesis of neuroinflammation by exacerbating harmful inflammatory responses and injuries. Therefore, our work provides a new theoretical basis for the occurrence of neuroinflammation and a new target for clinical drug development.
Materials and Methods
Cell Culture and Processing
HT22 mouse hippocampal neuronal cells and HEK-293T human embryonic kidney cells purchased from ATCC were cultured in Dulbecco’s modified Eagle’s medium (C11995500BT; Gibco; USA) emented with 10% fetal bovine serum (11,011–8611; Sijiqing; China) and 1% penicillin and streptomycin (15,070,063; Gibco; USA). To induce a cellular inflammatory response, the cells were treated with 1 μg/mL LPS for 24 hours. In addition, HT22 cells were treated with the transcription factor c-Fos/AP-1 inhibitor T-5224 (1 μM), the SOS1 inhibitor BAY293 (2 μM), the transcription factor AP-1 inhibitor SR11302 (2 μM), or the KRAS agonist KRAS-533 (10 μM) for 24 h, after which LPS was added and RT-real-time PCR, Western blotting and other experiments were performed. All cells were cultured at 37°C in a constant temperature incubator containing 5% CO2.
Plasmid Transfection
By RT-PCR method to amplify the FBL gene coding sequence primers sequence is: positive 5’ - CACACTGGACTAGTGGATCCCGCCACCATGAAGCCAGGTTTCAGCCC – 3’; reverse 5’- AGTCACTTAAGCTTGGTACCGAGTTCTTCACCTTGGGAGGTGGCCTGTAC – 3’. The PCR fragment and GV657 vector (CMV enhancer-MCS-3flag-polyA-EF1A-zsGreen-sv40-puromycin) were digested with BamHI/KpnI and ligated with T4 DNA ligase, CMV enhancer-FBL-3flag-polyA-EF1A-zsGreen-sv40-puromycin was generated.The wild-type GV657 vector was used as a control for FBL overexpression. The FBL short interfering RNA (siRNA) sequence was inserted into the GV298 vector (U6-MCS-Ubiquitin-Cherry-IRES-puromycin) with the sequence 5’ -CCACACAAATACCGCATGCTT-3 ‘, and the control siRNA sequence was inserted with the sequence 5’ -TTCTCCGAACGTGTCACGT-3 ‘. All plasmids mentioned above were purchased from Shanghai GenChem Technology Co., LTD (Shanghai, China). A total of 2×105 HEK-293T cells were inoculated in each well of a six-well plate, and when the cell density reached approximately 80%, the cells in each well were transfected with 2.5 μg of plasmid mixed with Lipofectamine™ 3000 in Opti-MEM according to the manufacturer’s instructions (A4124801; Thermo Fisher Scientific; USA). The cells were harvested 48 hours after transfection, and duplicate wells treated in parallel were used for the treatment experiment, Western blotting or RT-real-time PCR.
Protein Preparation and Western Blotting
The cells were collected and washed three times with precooled PBS, RIPA buffer supplemented with protease inhibitors and phosphatase inhibitors was added, and the cells were left on ice for 30 min. Afterward, the cells were collected with a spatula, the samples were centrifuged at 12,000 rpm for 30 min at 4°C, and the supernatant was collected. The protein concentration was quantified using a Pierce™ BCA Protein Assay Kit (23,227; Thermo Fisher Scientific; USA). Proteins (10–30 μg) were separated on a 10–12% SDS‒polyacrylamide gel by electrophoresis and then transferred to polyvinylidene difluoride (PVDF) membranes (IPVH00010; Millipore, USA). The PVDF membranes were blocked with 5% skim milk powder for 2 hours. The antibodies used in this study were rabbit anti-FBL (ABclonal, A0850, China, 1:100, 37 kDa) and rabbit anti-β-tubulin (BOSTER, Clone#17T15, China, 1:1000, 55 kDa). The membranes were incubated with primary antibody overnight at 4°C and then washed and incubated with horseradish peroxidase-labeled secondary antibody for 1 h at room temperature. After rinsing, the PVDF membranes were placed in a Bio-Rad ChemiDoc™ XRS system for scanning, and then Immobilon Western Chemiluminescent HRP Substrate (WBKLS0100; Millipore; USA) was added to visualize the signals. Densitometric analysis was performed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Real-Time RT-PCR
For RNA extraction, the culture medium was first removed, the cells were gently washed with PBS 3 times, and then the PBS was removed. The cell culture plate was placed on ice throughout the entire process. The cells were lysed by adding 1 mL of TRIzol to each well of a six-well plate, scraped off with a cell scraper, and aspirated into a 1.5 mL sterilized EP tube using a pipette gun. Afterward, 200 μL of chloroform was added for extraction, and the samples were vigorously shaken for 15s, left for 2 min, and centrifuged at 12,000 rpm for 15 min at 4°C. After centrifugation, 500 μL of isopropanol was added to each new EP tube, which was subsequently labeled. The supernatant was transferred to the appropriate EP tube, and the samples were mixed, left at room temperature for 30 min, and centrifuged at 12,000 rpm for 15 min at 4°C. The supernatant was decanted, 1 mL of 75% alcohol, and the samples were centrifuged at 10,000 rpm for 10 min at 4°C. The supernatant was poured off, the EP tube was inverted on filter paper to allow the ethanol to evaporate, and 30 μL of DEPC water was added. The mixture was then placed on ice, and the RNA concentration was measured. The reaction system for reverse transcription was prepared as follows: 1 μg of RNA, 4 μL of reverse transcriptase, and DEPC water to 20 μL. Reverse transcription was performed at 37°C for 15 min, 85°C for 5 s, and 4°C ∞. Total RNA was isolated from cells using TRIzol reagent (Invitrogen) and reverse transcribed into cDNA with a reverse transcription kit (RR037A; TaKaRa; Japan) according to the manufacturer’s instructions. The cDNA concentration was quantified using a real-time PCR kit (RR064A; TaKaRa; Japan), and real-time PCR was performed using a CFX Duet Real-Time PCR System. Fluorescence-based quantitative PCR was performed in triplicate using a CFX Duet Real-Time PCR System. The RNA expression levels of the target genes were calculated by the 2-ΔΔCt method, and GAPDH was used as an endogenous control. The PCR conditions for the experiments were 95°C for 30s, 55°C for 30s, and 72°C for 30s. Forty cycles were performed. All the experiments were performed three times independently. The sequences of the primers are listed in Table S1.
Immunofluorescence
A total of 2500 HT22 cells were inoculated in 15 mm confocal dishes, and after drug treatment, the cells were washed in PBS and fixed with 4% paraformaldehyde for 15 min, after which they were washed three times in PBS, permeabilized with 0.1% Triton X-100 on ice for 15 min, washed three times in PBS and then blocked in 5% BSA for 30 min. Finally, the cells were incubated with an anti-rabbit FBL antibody (ABclonal, A0850, China, 1:50) overnight at 4°C. The cells were incubated with secondary goat anti-rabbit IgG H&L (Alexa Fluor® 488-conjugated) (Abcam, ab150077, UK, 1:300) for 1 h. The cell nuclei were stained with DAPI for 5 min. Next, the cells were imaged using CLSM (Leica TSC SP 8, Germany) at an excitation wavelength of 488 nm and an emission wavelength of 500 to 700 nm.
Cell Viability Assay
Cell viability was assessed with a CCK8 cell proliferation kit (C0038; Biyuntian; China). Briefly, the cells were inoculated at a density of 2000 cells/well and cultured in 96-well plates overnight. Different concentrations of LPS were added to each well for treatment (0, 50, 100, 150, or 200 μM), and the cells were incubated for 24, 48, or 72 hours. The plates were removed from the incubator at the indicated time, 10 μL of fresh medium and CCK8 reagent (100 μL of medium and 10 μL of CCK8 reagent) were added to each well, and the cells were incubated (at 37°C) for 1 h away from light. The absorbance of the cells at a wavelength of 450 nm was measured by an enzyme labeling machine. The cell proliferation rate was calculated formulas (Experiment A—Blank A) / (Control A—Blank A), and the half-maximal inhibitory concentration (IC50) was calculated using GraphPad Prism 9.0.
Apoptosis Assay
Cell activity and cytotoxicity were determined using the Calcein/PI Cell Activity and Cytotoxicity Assay Kit (C2015M; Biyuntian; China) according to the manufacturer’s instructions. Briefly, cells were inoculated in a six-well plate, the culture medium was discarded after treatment, and staining working solution was added with serum-free medium at a ratio of 1:100. Calcein AM was used to stain live cells and appeared ass green fluorescence, while propidium iodide was used to stain dead cells and appeared ass red fluorescence. After 30 min of staining, fluorescence microscopy was performed for detection and analysis.
RNA Sample Preparation and Sequencing
Three replicate samples of cells transfected with an FBL overexpression plasmid or control plasmid were collected. RNA extraction was performed using TRIzol reagent. To synthesize second-strand cDNA, the RNA samples were fragmented, and random hexamers were subsequently annealed to the fragments. To purify the cDNA fragments, end repair and adaptor ligator were performed, and then the QIAquick PCR extraction kit was used. The enzyme UNG was used to dissolve the second-strand cDNA. Ribosomal RNA (rRNA) was subsequently removed from the samples using the GenSeq® rRNA Removal Kit (GS-ET-001; GenSeq; China). After rRNA removal, the samples were used to construct sequencing libraries using the GenSeq® Low Input RNA Library Prep Kit (GS-ET-001; GenSeq; China) following the manufacturer’s instructions. The constructed sequencing libraries were subjected to quality control and quantification by a BioAnalyzer 2100 system (Agilent Technologies; USA), and 150 bp sequencing was subsequently performed using an Illumina NovaSeq 6000 instrument. RNA and library quality control consisted of three parts: First RNA quantification control, RNA concentration of each sample was measured using a NanoDrop ND-1000 instrument (Thermo Fisher Scientific, Waltham, MA, USA). The OD260/OD280 value was used as an indicator of RNA purity. If the OD260/OD280 value ranges from 1.8 to 2.1, the RNA purity is qualified, and the QC Results are qualified. A second RNA integrity quality control, RNA integrity and gDNA contamination were measured using denaturing agarose gel electrophoresis. The third RNA library quality control, library quality was checked using an Agilent 2100 Bioanalyzer instrument.
RNA-Seq Data Processing and Analysis
After sequencing on an Illumina NovaSeq 6000 sequencer, the reads were obtained. Quality control was performed using Q30, and high-quality reads were obtained by dejoining and removing low-quality reads using Cutadapt software (v1.9.3). The high-quality reads were compared to the reference genome using HISAT2 software (v2.0.4). Then, HTSeq software (v0.9.1) was used to obtain raw gene counts as mRNA expression profiles. Differentially expressed mRNAs were identified using edgeR software (v3.16.5) to normalize and calculate the fold change and p value between the two sets of samples. ∣log2FoldChange∣>1, p-value< = 0.05 as the threshold for difference screening. The heatmap 2 function of R was used to perform clustering analysis of differentially expressed mRNAs with standardized count number.GO and KEGG pathway enrichment analyses of the differentially expressed mRNAs were performed. P value < = 0.05 was used as the threshold for significant enrichment.
Molecular Docking Analysis
Molecular docking of the FBL gene to the AP-1 protein was performed. The 3D structure of the AP-1 protein was downloaded from the PDB database (RCSB PDB: Homepage). Water molecules and primitive ligands were removed from the target proteins by PyMOL, and then the target proteins were imported into AutoDock Tools 1.5.6 for hydrogenation, charge calculations, and nonpolar hydrogen binding. The results were subsequently stored in PDBQT format. The size of the grid box was set to 40×40 × 40. Finally, molecular docking analysis was performed with AutoDock Vina using the CMD command,18 and the results were visualized using PyMOL.
Bioinformatics Analysis
Principal component analysis (PCA) was used to assess the correlation between duplicates and differential gene expression. Audics was used to analyze differential gene expression, and the genes that were differentially expressed after DHA treatment were annotated with GO terms (http://www.geneontology.org/). Biological pathway enrichment analysis was performed by searching the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.ad.jp/kegg/).
Statistical Analysis
All experiments were performed with at least three independent biological replicates. Statistical analysis was performed using GraphPad Prism 9.0. All the data are expressed as the mean and standard error of the mean (SEM). Statistical analyses were performed using Student’s t–test for comparisons between two groups and one-way analysis of variance (ANOVA) for comparisons among multiple groups. p values <0.05 were considered to indicate statistical significance. The statistical parameters are detailed in the legend of each figure.
Results
FBL Expression is Elevated in an HT22 Cell Model of LPS-Induced Inflammation
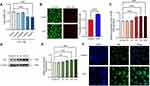
The inflammatory effects of LPS have been investigated in macrophages and microglia; in the present study, we extended our investigations to the HT22 mouse hippocampal neuronal cell line. To assess the effects of LPS on the viability and proliferation of HT22 cells, we incubated the cells with different doses of LPS (0, 0.5, 0.75, 1.0, or 1.5 μg/mL) for 24 h. The CCK-8 assay showed that LPS treatment significantly reduced the viability of HT22 cells in a dose-dependent manner (Figure 1A). Approximately 25% of the HT22 cells died after 24 h of treatment with 1.0 μg/mL LPS; therefore, 1.0 μg/mL LPS was used for subsequent experiments. The percentage of dead cells was then calculated using the Calcein/PI Cell Activity and Cytotoxicity Assay Kit, and in agreement with the CCK-8 assay results (Figure 1B), there were more dead cells in the LPS-treated group than in the blank group, suggesting that LPS caused a decrease in cell viability.
There is growing evidence that nucleoli play a role in regulating cellular processes related to health and disease and that FBL knockdown prior to infection increases intracellular bacterial clearance, reduces inflammation, and enhances cell survival. To determine the effect of the LPS treatment time on FBL expression levels, RT-real-time PCR and Western blotting were performed, and the results showed that FBL mRNA and protein expression levels in HT22 cells increased with increasing LPS treatment time, were significantly greater after 6 h of LPS treatment and reached their highest levels at 24 h of LPS treatment (Figure 1C, Figure 1D, Figure 1E). These results suggest that the LPS-induced decrease in HT22 cell viability may be associated with elevated expression of FBL, which is involved in the early processing of rRNA, specifically the 5′ external transcribed spacer (5′ ETS), and is localized in the dense protofibrillar component (DFC) of the nucleolus. An increased fluorescence intensity of FBL (green) was observed in the nucleus after 24 h of LPS treatment using immunofluorescence techniques (Figure 1F), further verifying the elevated expression of FBL after LPS treatment. These results indicate that FBL is mainly localized in the nucleus of cells, that the expression of FBL is greater in the nucleus of cells treated with LPS, and that the expression level of FBL in the nucleus of cells is positively correlated with inflammation.
FBL Knockdown Has a Protective Effect Against Inflammation in Cell Models
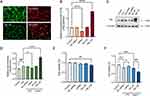
To investigate the effect of FBL on LPS-induced inflammation in a cell model, we modulated FBL expression by plasmid transfection to investigate the possible mechanisms by which FBL affects cellular inflammatory responses. We transfected FBL knockdown (U6-MCS-Ubiquitin-Cherry-IRES-puromycin) and overexpression (CMV enhancer-MCS-3flag-polyA-EF1A-zsGreen-sv40-puromycin) plasmids into HEK 293T cells, and then fluorescence microscopy was performed. The transfection efficiency of the FBL overexpression plasmid, which contained green fluorescent protein (GFP), was assessed at an excitation wavelength of 488 nm, and it was found to be 90%; the transfection efficiency of the FBL knockdown plasmid, which contained the red fluorescent protein Cherry, was evaluated at an excitation wavelength of 587 nm and was found to be 90%. Thus, the results indicated that the plasmids were successfully transfected into the HEK 293T cells (Figure 2A). The effect of plasmid-mediated FBL knockdown and overexpression was further verified using qPCR, which showed that FBL gene expression in the FBL-transfected groups significantly differed from that in the control and NC groups (Figure 2B), indicating the plasmid transfection successful modulated FBL gene expression.
In addition, the expression of FBL in an HEK-293T cell model of inflammation simulated by LPS was subsequently examined to investigate the possible mechanism of cellular inflammatory damage caused by FBL. The results showed increased FBL protein expression in the inflammatory environment of LPS-treated cells, and FBL knockdown and overexpression plasmid transfection followed by treatment with LPS significantly altered the expression of FBL (Figure 2C, Figure 2D). To further investigate the effect of FBL on cell viability, the CCK-8 assay was performed, and the results showed that when HEK-293T cells were transfected with the FBL overexpression plasmid, cell viability decreased compared with that in the control group; however, knockdown of FBL (induced by transfection of the siRNA plasmid) resulted in no significant difference in cell viability compared with the control group. (Figure 2E). The CCK-8 assay results showed that the viability of cells transfected with the FBL overexpression plasmid and treated with LPS decreased; however, cell viability recovered to a certain degree when FBL was knocked down (Figure 2F). The above results suggest that FBL overexpression promotes apoptosis and that FBL knockdown improves survival during LPS-induced inflammation.
FBL is Regulated by the Upstream RAS Signaling Pathway
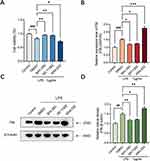
Previous findings have shown that LPS induces the expression of NF-kB and RAS/MAPK pathway components as well as proinflammatory mediators in cells, so we hypothesized that overexpression of FBL promotes apoptosis through the classical MAPK signaling pathway. To explore the specific mechanism by which FBL regulates apoptosis, several key target molecules in the MAPK signaling pathway were activated or inhibited by using the SOS1 inhibitor BAY-293, the AP-1 inhibitor SR 11302, and the KRAS agonist KRA-533 in vitro. HT22 cells were treated with the agonist and inhibitors of these three key targets of the MAPK signaling pathway for 24 h, followed by LPS treatment for 24 h. The CCK-8 assay results showed that LPS treatment significantly reduced cell viability, and the inhibitors effectively restored cell viability; in contrast, the agonist aggravated cell apoptosis to a certain extent (Figure 3A). qPCR showed that both SR11302 and BAY-293 significantly inhibited the expression of FBL at the mRNA level compared with that in control cells, and the agonist KRAS significantly increased the mRNA level of FBL (Figure 3B). Western blot analysis revealed that BAY-293 and SR11302 significantly reduced the expression level of FBL in the LPS-treated cell model, and the agonist KRAS significantly increased the expression level of FBL (Figure 3C, Figure 3D). As expected, the agonist promoted FBL production, leading to apoptosis, and treatment with the inhibitors blocked this effect. These results suggest that the proinflammatory effects of LPS may be related to the enhancement of FBL expression via regulation of the RAS/MAPK pathway in activated HT22 cells and that the inhibition of c-Jun/c-Fos/FBL signaling can effectively reduce inflammatory symptoms.
AP-1 is an intracellular transcriptional activator and a heterodimer composed of c-Fos and c-Jun. AP-1 heterodimerization is mediated by a leucine zipper, and the AP-1 heterodimer binds to genes through specific conserved sequences to initiate gene expression. The structure of the AP-1 protein was obtained from the PDB database, and the structures of the AP-1 protein and the FBL gene were imported into AutoDock Tools 1.5.6. The molecular docking results showed that a hydrophilic amino acid site of the AP-1 protein that is rich in arginine and aspartic acid binds to the promoter of the FBL gene (>NC_000019.10:39,847,389–39,847,395 Homo sapiens chromosome 19, GRCh38.p13 Primary Assembly, 5′-TGAATCA-3′), indicating that FBL is regulated at the transcriptional level by the AP-1 complex downstream of Ras signaling (Figure 4A).
To verify this prediction, FBL expression was measured in cells pretreated with the c-Fos/AP-1 inhibitor T-5224 for 24 h and stimulated with LPS for 24 h. Treatment with the inhibitor T-5224 significantly reduced FBL protein and mRNA levels after LPS stimulation (Figure 4B, Figure 4C, Figure 4D). The above results indicate that T-5224 inhibits the LPS-induced activation of RAS/MAPK in HT22 cells and that FBL expression is regulated by the upstream molecules SOS1, AP-1 and KRAS.
FBL Regulates the NF-kB Signaling Pathway to Promote Apoptosis
The above studies demonstrated that AP-1 family members such as c-Jun and c-Fos activate FBL and increase inflammatory responses in HT22 cells. To explore the specific molecular regulatory mechanisms downstream of FBL-triggered cellular inflammation, we performed transcriptome sequencing of cells transfected with FBL overexpression plasmids. We comprehensively examined the transcriptomes of the samples using an Illumina NovaSeq sequencer and performed transcriptomic studies to determine the molecular mechanisms by which FBL promotes HT22 cell injury. To determine the consistency of the samples, correlation analysis was used, and the Pearson correlation coefficients (PCCs) showed strong intragroup correlations and weak intergroup correlations (Figure 5A).
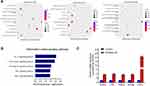
Transcriptome sequencing of FBL-overexpressing HT22 cells revealed a total of 5375 differentially expressed genes. Among them, 3225 genes were upregulated and 2362 genes were downregulated (Figure 5B), with 558 genes being specifically regulated by FBL (Figure 5C). The differentially expressed genes between the FBL-overexpressing group and the control group were clustered, and the 10 upregulated genes are displayed in a heatmap (Figure 5D).
The differentially expressed mRNAs were subjected to GO functional analysis to determine the functions of these mRNAs. The results showed that the upregulated mRNAs in the FBL-overexpressing group compared to the control group were enriched mainly in transcriptional regulatory complexes and nucleosome regions. Regarding biological processes, FBL was found to regulate signal transduction and genes related to the positive regulation of biological processes. Regarding molecular functions, FBL was found to mainly regulate genes related to receptor binding, DNA transcription factor activity, and RNA polymerase activity (Figure 6A). The results of KEGG pathway analysis indicated that the differentially expressed genes were enriched in the following five signaling pathways associated with inflammatory responses: (1) the IL-17 signaling pathway; (2) the T-cell receptor signaling pathway; (3) the NF-kappa B signaling pathway; (4) the TNF signaling pathway; and (5) the Rap1 signaling pathway (Figure 6B). This indicated that FBL acts mainly through these pathways in HT22 cells.
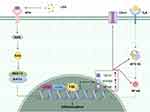
In this study, the NF-kB signaling pathway was found to be associated with LPS-induced inflammation, and several key molecules in this signaling pathway were shown to be significantly upregulated. Some of the identified differentially expressed genes, specifically, CD14, MYD88, TNF, TRADD, and NFKB1, are involved in the NF-kB signaling pathway and were selected for validation by real-time RT-PCR; we found that all five molecules were differentially expressed (Figure 6C). In this study, it was verified that FBL is regulated upstream by the RAS/MAPK pathway and that the upregulation of FBL can in turn promote the release of relevant inflammatory factors in the NF-kB signaling pathway, further promoting the generation of inflammation and apoptosis in HT22 cells (Figure 7).
Discussion
Our study showed that FBL expression was activated through the MAPK/RAS signaling pathway in an LPS-induced neuroinflammation model and that the downstream activation of FBL through the NF-kB signaling pathway further promoted inflammation and apoptosis. In addition, inhibitors of several key targets of the RAS signaling pathway (BAY-293, SR11302, and T-5224) inhibited the expression of FBL, thereby reducing inflammation and apoptosis. These results may provide new insights into the mechanisms of neuroinflammation (Figure 7).
Neuroinflammation is an immune response activated by glial cells in the CNS that usually occurs in response to nerve injury, infection, toxins and other stimuli or in the context of autoimmunity.19 Neuroinflammation is closely related to the progression of neurodegenerative diseases such as AD, PD, ALS and MS.20 Among the different cell types involved in neuroinflammation, astrocytes have emerged as key players due to their multiple functions in the CNS.21 Astrocytes play dual roles in neuroinflammation. On the one hand, they secrete anti-inflammatory factors to promote tissue repair and nerve recovery. On the other hand, they can produce proinflammatory cytokines, reactive oxygen species, and other neurotoxic substances, which can exacerbate the neuroinflammatory response and lead to neurodegeneration. These inflammatory mediators interact with several biomolecules, such as membrane lipids, proteins, and nucleic acids, to further impair neuronal function by altering synaptic, myelin, and cell body structure. Neuroinflammation is characterized by the release of proinflammatory cytokines from the CNS and activation of the innate immune response.22,23 Preclinical studies have shown that anti-inflammatory drugs exert therapeutic effects, improving the prognosis of neuroinflammatory disorders, but reports from clinical trials are less promising.24 Therefore, there is a need for new therapeutic agents aimed at restoring the balance between proinflammatory and anti-inflammatory processes to alleviate neuroinflammation.
FBL is a 2’-O-methyltransferase that is mainly localized in the nucleolus, the region where RNA transcription and pre-RNA processing occur. FBL forms complexes with several structural proteins and box C/D (mall nucleolar ribonucleic acids (snoRNAs) and plays important roles in pre-rRNA processing, methylation, and ribosome assembly. The complexes formed by FBL can catalyze the specific 2’-O-ribose methylation of rRNA sequences.25 The sequence, structure and function of FBL are highly conserved. FBL gene expression plays an important role in early embryonic development. The FBL homolog in yeast, Nop1p, is also localized in the nucleolus and is involved in 18S rRNA processing and maturation.26 Serum from patients with scleroderma, an autoimmune disease, often contains high levels of antibodies against FBL.27 The exact mechanism by which FBL affects these processes has not been discovered, but some experimental data suggest that upregulation or downregulation of FBL can alter ribosomal structure and thus the relative translation efficiency of various mRNAs, potential contributing to the regulation of cell proliferation, cancer progression, and aging.28 Varnesh Tiku et al revealed an evolutionarily conserved novel function of FBL in the regulation of mammalian innate immunity.29 FBL knockdown was found to increase intracellular bacterial clearance, alleviate inflammation, and increase cell survival. In the present study, we evaluated the regulation of FBL expression by relevant signaling components in LPS-stimulated HT22 cells. Our results confirmed that HT22 cells exhibit a significant increase in FBL expression upon LPS stimulation and that FBL promotes inflammation and apoptosis.
Studies have shown that LPS can induce inflammatory responses and free radical production in animals and renal cells and that RAS expression and activation are markedly increased under various inflammatory conditions in mice and humans. The molecular and pharmacological blockade of RAS inhibits inflammatory activity in vitro and in vivo by inhibiting the AP-1 pathway in immune cells and cancer cells.30 In the present study, we confirmed that LPS promotes inflammation by increasing FBL expression through the RAS signaling pathway. RAS superfamily GTPases play important roles in matrix and immune cell activation during inflammatory responses.31 The Ras protein has a binding site for GTP or GDP and can control cell signal transduction by transforming between two conformations, thereby regulating cell differentiation, proliferation and apoptosis. In cells in the resting state, growth factor receptor-bound protein 2 (Grb2) binds to son of sevenless homolog (SOS) to form a complex in the cytoplasm. When receptor tyrosine kinase (RTK) receptors are activated, the Grb2-SOS complex binds to these receptors and transports them to the cell membrane to recruit SOS, thereby increasing the SOS concentration at the plasma membrane. SOS has nucleotide transferase activity and activates the downstream target protein Ras protein for signal transduction.32 In previous studies, the major transcription factors regulating proinflammatory mediators were studied, and molecular docking analysis was used to determine that AP-1 interacts with FBL. We hypothesize that an increase in the concentration of SOS at the plasma membrane and activation of Ras proteins in HT22 cells upon exogenous LPS stimulation further facilitates the interaction of the AP-1 complex with FBL and promotes FBL expression (Figure 7). Moreover, an AP-1 inhibitor, SOS1 inhibitor and KRAS agonist could regulate the expression of FBL, proving that FBL is regulated by upstream RAS signaling. Our findings suggest that RAS signaling occurs upstream of FBL, providing not only additional information about the role of the pathway downstream of RAS signaling in the regulation of neuroinflammation but also a potential mechanism for the transcriptional regulation of FBL.
To further explore the specific mechanism by which FBL promotes inflammation, transcriptomic sequencing was performed, and the results showed that FBL activated the NF-κB signaling pathway and upregulated inflammation-related genes (CD14, MYD88, TNF, TRADD, and NFKB1). Specifically, FBL upregulation significantly activated cluster of differentiation 14 (CD14), formerly known as the MY23 antigen, a surface antigen in the NF-kB signaling pathway that is a member of the cell surface glycoprotein family. CD14 is a high-affinity receptor for the LPS-lipopolysaccharide binding protein (LBP) complex in gram-negative bacteria. CD14 recognizes and binds to LPS, induces tyrosine phosphorylation, activates intracellular myeloid differentiation factor 88 (MYD88), and then activates NF-kB through gradual phosphorylation, inducing the translocation of NF-kB from the cytoplasm into the nucleus, initiating the transcription of related cytokine genes, and completing the NF-kB pathway.33 NF-kB regulatory genes encode mainly cytokines involved in the immune response and inflammatory response, immune receptor molecules and acute phase proteins. CD14 plays an important role in a series of reactions induced by the body’s immune and defense systems. Therefore, the CD14 molecule and its signaling pathway play important roles in the immune response and inflammatory response in the body. The NF-kB pathway ultimately leads to the release of IL-1, IL-6, TNF-α, NO, etc.,34 leading to inflammation and other diseases (Figure 7). Our results provide a theoretical basis for the downstream mechanism by which FBL regulates inflammation via modulation of the NF-kB signaling pathway.
Although FBL expression was analyzed by Western blotting, RT-real-time PCR, and immunofluorescence, it is important to consider incorporating other relevant assessment methods in future studies to further confirm the reliability of our results. In addition to assessing mRNA and protein expression, quantitative measurements of FBL enzyme activity can be performed using fluorescence assays.35 Furthermore, epigenetic approaches can be conducted to assess the ability of FBL to regulate the methylation of its downstream targets. It is also worthwhile to investigate the effect of LPS on FBL transcriptional activity using reporter gene analysis. The limitation of this study is the lack of genetic inhibition studies to validate our findings; although the importance of c-FOS, AP-1 and SOS1 in LPS-induced FBL expression in HT22 cells was demonstrated by using specific inhibitors, the expression of c-FOS, AP-1 and SOS1 in HT22 cells was not downregulated by shRNA, and plasmids need to be constructed for further validation. Another limitation is that this study involved only in vitro experiments, and an in vivo animal model is needed to further clarify the regulatory role of FBL in neuroinflammation in vivo and the related mechanism. Despite these limitations, this study provides a new theoretical basis for the neuronal sensing of inflammatory stimuli and identifies relevant intervention targets and regulatory pathways of the inflammatory response. In combination with existing research, these results are expected to provide new ideas for research on and the clinical treatment of neuroinflammation.
Conclusion
In summary, we have conducted a comprehensive study of the functional and regulatory mechanisms of LPS-mediated FBL expression in a mouse hippocampal neuronal cell line. The activation of the MAPK/RAS signaling pathway was found to promote the expression of FBL after LPS stimulation in HT22 cells, and FBL was shown to further activate the downstream NF-kB signaling pathway to promote cellular inflammation and apoptosis. In addition, our study revealed a novel mechanism by which FBL inhibition protects against neuroinflammation, providing potential new targets for the study of neuroinflammation. In the future, the mechanism of FBL in neuroinflammation still needs to be further explored.
Abbreviations
LPS, lipopolysaccharide; CNS, central nervous system; NF-kB, nuclear factor kB; AP-1, activating protein-1; NFAT, nuclear factor of activated T cell; STAT, signal transducer and activator of transcription; MAPKs, mitogen-activated protein kinases; ERK, extracellular signal-regulated kinase; JNK, c-Jun N-terminal kinase; NLRP3, NLR family pyrin domain containing 3; FBL, Fibrillarin; snoRNP, small nucleolar ribonucleoprotein; snoRNAs, small nucleolar ribonucleic acids; NFTs, neurofibrillary tangles; NMDA, N-methyl-D-aspartic acid; Aβ, amyloid plaques; SOS1, son of sevenless homolog 1; RTKs, receptor tyrosine kinases; Grb2, growth factor receptor-bound protein 2; LBP, lipopolysaccharide-binding protein; MYD88, myeloid differentiation factor 88; CD14, cluster of differentiation 14; IL, interleukin; TNF-α, tumor necrosis factor α; CSF, cerebrospinal fluid; real-time RT-PCR, real-time reverse transcription-polymerase chain reaction; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Data Sharing Statement
The data used in this study are available from the corresponding author upon reasonable request.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 82171321, 82171363, and 82371381).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Tian H, Wang K, Jin M, Li J, Yu Y. Proinflammation effect of Mst1 promotes BV-2 cell death via augmenting Drp1-mediated mitochondrial fragmentation and activating the JNK pathway. J Cell Physiol. 2020;235(2):1504–1514. doi:10.1002/jcp.29070
2. Pan S, Lv Z, Wang R, et al. Sepsis-induced brain dysfunction: pathogenesis, diagnosis, and treatment. Oxid Med Cell Longev. 2022;2022:1328729. doi:10.1155/2022/1328729
3. Broos JY, van der Burgt RTM, Konings J, et al. Arachidonic acid-derived lipid mediators in multiple sclerosis pathogenesis: fueling or dampening disease progression? J Neuroinflammation. 2024;21(1):21. doi:10.1186/s12974-023-02981-w
4. So YJ, Lee JU, Yang GS, et al. The potentiality of natural products and herbal medicine as novel medications for parkinson’s disease: a promising therapeutic approach. Int J Mol Sci. 2024;25(2):1071. doi:10.3390/ijms25021071
5. Gaikwad S, Senapati S, Haque MA, Kayed R. Senescence, brain inflammation, and oligomeric tau drive cognitive decline in Alzheimer’s disease: evidence from clinical and preclinical studies. Alzheimer’s Dementia. 2024;20(1):709–727. doi:10.1002/alz.13490
6. Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls toll-like receptor signaling. Cell. 2006;125(5):943–955. doi:10.1016/j.cell.2006.03.047
7. Wu H, Zheng J, Xu S, et al. Mer regulates microglial/macrophage M1/M2 polarization and alleviates neuroinflammation following traumatic brain injury. J Neuroinflammation. 2021;18(1):2. doi:10.1186/s12974-020-02041-7
8. Alzahrani NA, Bahaidrah KA, Mansouri RA, Alsufiani HM, Alghamdi BS. Investigation of the optimal dose for experimental lipopolysaccharide-induced recognition memory impairment: behavioral and histological studies. J Integra Neurosci. 2022;21(2):49. doi:10.31083/j.jin2102049
9. Barnabei L, Laplantine E, Mbongo W, Rieux-Laucat F, Weil R. NF-κB: at the borders of autoimmunity and inflammation. Front Immunol. 2021;12:716469. doi:10.3389/fimmu.2021.716469
10. Wan P, Zhang S, Ruan Z, et al. AP-1 signaling pathway promotes pro-IL-1β transcription to facilitate NLRP3 inflammasome activation upon influenza A virus infection. Virulence. 2022;13(1):502–513. doi:10.1080/21505594.2022.2040188
11. Lee HG, Kim LK, Choi JM. NFAT-Specific Inhibition by dNP2-VIVITAmeliorates autoimmune encephalomyelitisby regulation of Th1 and Th17. Mol Ther Meth Clin Develop. 2020;16:32–41.
12. Jain M, Singh MK, Shyam H, et al. Role of JAK/STAT in the neuroinflammation and its association with neurological disorders. Annal Neurosci. 2021;28(3–4):191–200. doi:10.1177/09727531211070532
13. Zgórzyńska E, Stulczewski D, Dziedzic B, Su KP, Walczewska A. Docosahexaenoic fatty acid reduces the pro-inflammatory response induced by IL-1β in astrocytes through inhibition of NF-κB and AP-1 transcription factor activation. BMC neuro. 2021;22(1):4. doi:10.1186/s12868-021-00611-w
14. Dresselhaus EC, Meffert MK. Cellular specificity of NF-κB function in the nervous system. Front Immunol. 2019;10:1043. doi:10.3389/fimmu.2019.01043
15. Cai L, Gong Q, Qi L, et al. ACT001 attenuates microglia-mediated neuroinflammation after traumatic brain injury via inhibiting AKT/NFκB/NLRP3 pathway. Cell Commun Signaling. 2022;20(1):56. doi:10.1186/s12964-022-00862-y
16. Yi Y, Li Y, Meng Q, et al. A PRC2-independent function for EZH2 in regulating rRNA 2’-O methylation and IRES-dependent translation. Nat Cell Biol. 2021;23(4):341–354. doi:10.1038/s41556-021-00653-6
17. Iyer-Bierhoff A, Krogh N, Tessarz P, Ruppert T, Nielsen H, Grummt I. SIRT7-dependent deacetylation of fibrillarin controls histone H2A Methylation and rRNA synthesis during the cell cycle. Cell Rep. 2018;25(11):2946–2954.e2945. doi:10.1016/j.celrep.2018.11.051
18. Xue Q, Liu X, Russell P, et al. Evaluation of the binding performance of flavonoids to estrogen receptor alpha by autodock, autodock vina and surflex-dock. Ecotoxicol Environ Saf. 2022;233:113323. doi:10.1016/j.ecoenv.2022.113323
19. Xu X, Gao W, Cheng S, et al. Anti-inflammatory and immunomodulatory mechanisms of atorvastatin in a murine model of traumatic brain injury. J Neuroinflammation. 2017;14(1):167. doi:10.1186/s12974-017-0934-2
20. Singh J, Habean ML, Panicker N. Inflammasome assembly in neurodegenerative diseases. Trends Neurosci. 2023;46(10):814–831. doi:10.1016/j.tins.2023.07.009
21. Desai A, Park T, Barnes J, Kevala K, Chen H, Kim HY. Reduced acute neuroinflammation and improved functional recovery after traumatic brain injury by α-linolenic acid supplementation in mice. J Neuroinflammation. 2016;13(1):253. doi:10.1186/s12974-016-0714-4
22. Capizzi A, Woo J, Verduzco-Gutierrez M. Traumatic brain injury: an overview of epidemiology, pathophysiology, and medical management. Med Clin North Am. 2020;104(2):213–238. doi:10.1016/j.mcna.2019.11.001
23. Shively SB, Priemer DS, Stein MB, Perl DP. Pathophysiology of traumatic brain injury, chronic traumatic encephalopathy, and neuropsychiatric clinical expression. Psychiatric Clini North Amer. 2021;44(3):443–458. doi:10.1016/j.psc.2021.04.003
24. Loane DJ, Kumar A. Microglia in the TBI brain: the good, the bad, and the dysregulated. Exp Neurol. 2016;275 Pt 3(0 3):316–327. doi:10.1016/j.expneurol.2015.08.018
25. Barros-Silva D, Klavert J, Jenster G, Jerónimo C, Lafontaine DLJ, Martens-Uzunova ES. The role of oncoSnoRNAs and ribosomal RNA 2’-O-methylation in cancer. RNA Biology. 2021;18(sup1):61–74. doi:10.1080/15476286.2021.1991167
26. Rothé B, Manival X, Rolland N, et al. Implication of the box C/D snoRNP assembly factor Rsa1p in U3 snoRNP assembly. Nucleic Acids Res. 2017;45(12):7455–7473. doi:10.1093/nar/gkx424
27. Keppeke GD, Satoh M, Kayser C, et al. A cell-based assay for detection of anti-fibrillarin autoantibodies with performance equivalent to immunoprecipitation. Front Immunol. 2022;13:1011110. doi:10.3389/fimmu.2022.1011110
28. Shubina MY, Musinova YR, Sheval EV. Proliferation, cancer, and aging-novel functions of the nucleolar methyltransferase fibrillarin? Cell Biol Int. 2018;42(11):1463–1466. doi:10.1002/cbin.11044
29. Tiku V, Kew C, Mehrotra P, Ganesan R, Robinson N, Antebi A. Nucleolar fibrillarin is an evolutionarily conserved regulator of bacterial pathogen resistance. Nat Commun. 2018;9(1):3607. doi:10.1038/s41467-018-06051-1
30. Yang WS, Kim HG, Kim E, et al. Isoprenylcysteine carboxyl methyltransferase and its substrate ras are critical players regulating TLR-mediated inflammatory responses. Cells. 2020;9(5):1216. doi:10.3390/cells9051216
31. Arrazola Sastre A, Luque Montoro M, Gálvez-Martín P, et al. Small GTPases of the ras and rho families switch on/off signaling pathways in neurodegenerative diseases. Int J Mol Sci. 2020;21(17):6312. doi:10.3390/ijms21176312
32. Lin CW, Nocka LM, Stinger BL, et al. A two-component protein condensate of the EGFR cytoplasmic tail and Grb2 regulates ras activation by SOS at the membrane. Proc Natl Acad Sci USA. 2022;119(19):e2122531119. doi:10.1073/pnas.2122531119
33. Ciesielska A, Matyjek M, Kwiatkowska K. TLR4 and CD14 trafficking and its influence on LPS-induced pro-inflammatory signaling. Cell Mol Life Sci. 2021;78(4):1233–1261.
34. Fu YJ, Xu B, Huang SW, et al. Baicalin prevents LPS-induced activation of TLR4/NF-κB p65 pathway and inflammation in mice via inhibiting the expression of CD14. Acta Pharmacol Sin. 2021;42(1):88–96. doi:10.1038/s41401-020-0411-9
35. Yang T, Low JJA, Woon ECY. A general strategy exploiting m5C duplex-remodelling effect for selective detection of RNA and DNA m5C methyltransferase activity in cells. Nucleic Acids Res. 2020;48(1):e5. doi:10.1093/nar/gkz1047
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.