Back to Journals » International Journal of General Medicine » Volume 14
Extracellular Matrix-Associated Pathways Promote the Progression of Gastric Cancer by Impacting the Dendritic Cell Axis
Authors Wang Z, Wang Z, Hu X, Han Q, Chen K, Pang G
Received 14 August 2021
Accepted for publication 29 September 2021
Published 13 October 2021 Volume 2021:14 Pages 6725—6739
DOI https://doi.org/10.2147/IJGM.S334245
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Zhenlin Wang, 1,* Zunyun Wang, 2,* Xianyu Hu, 1 Qijun Han, 1 Ke Chen, 1 Gang Pang 2
1Department of General Surgery, The First Affiliated Hospital of Anhui Medical University, Hefei, 230022, Anhui, People’s Republic of China; 2Department of Human Anatomy, School of Basic Medicine Sciences, Anhui Medical University, Hefei, 230032, Anhui, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ke Chen
Department of General Surgery, The First Affiliated Hospital of Anhui Medical University, 218th Jixi Road, Hefei, 230022, Anhui, People’s Republic of China
Email [email protected]
Gang Pang
Department of Human Anatomy, School of Basic Medicine Sciences, Anhui Medical University, 69th Meishan Road, Hefei, 230032, People’s Republic of China
Email [email protected]
Background: Gastric cancer (GC) is the third most frequent malignant tumour in the Chinese population, let alone the whole world. Recently, most prognostic models have only focused on the levels of several genes, miRNAs, lncRNAs, gene mutations, or DNA methylation; however, the activation status of biological pathways is more stable and can reflect the comprehensive inner conditions of tumours.
Methods: We collected samples from the Cancer Genome Atlas Stomach Adenocarcinoma (TCGA-STAD) cohort and GSE62254 cohort, with a total of 594 patients. We employed GSEA to first compare the diverse activated signalling pathways between dead GC patients and living patients. The least absolute shrinkage and selection operator (LASSO) regression analysis was subsequently performed by the “glmnet” package to generate a prognostic signature.
Results: We extracted a total of 218 genes from the KEGG Focal Adhesion and KEGG ECM Receptor Interaction pathways, which showed significant activation in dead GC patients in two enrolled cohorts, for subsequent LASSO analysis. In the TCGA-STAD cohort, patients in the high-risk group faced a significantly poorer prognosis than those in the low-risk group (P < 0.001, HR: 4.62, 95% CI: 3.447– 6.183), with an AUC of 0.694. In the GSE62254 cohort, the HR value was 4.94 (95% CI: 3.413– 7.165), and the AUC value was as high as 0.834. A high-risk score and poor prognosis correlated with infiltrated dendritic cells, and the receptor of IFN-α was also positively linked with the risk score, as well as poor prognosis. GC patients with high-risk scores were more likely to respond to CTLA4 treatment but not PD1 treatment.
Conclusion: Taken together, we established and verified an extracellular matrix prognostic model of gastric cancer patients. The model can be used to evaluate the risk of death of GC patients, as well as the response to anti-CTLA4 immunotherapy.
Keywords: gastric cancer, extracellular matrix, dendritic cell, immunotherapy, prognosis
Corrigendum for this paper has been published
Introduction
In summary, gastric cancer (GC) is the 6th leading cause of cancer morbidity and the 5th leading cause of cancer-specific mortality.1 Additionally, GC is the third most frequent malignant tumour in Chinese men and fourth most frequent in Chinese women in recent decades.2 The clinical symptoms of GC are frequently confused with benign gastric disease, including indigestion, heartburn, loss of appetite, and bloating of the stomach after meals; therefore, more than 60% of GC patients are diagnosed only at an advanced tumour stage, which limits clinical treatment.3 Clinical treatment with surgery, radiotherapy, or chemotherapy cannot provide satisfactory effectiveness for GC patients, and the 5-year survival rate of GC is less than 30% due to advanced metastatic tumours or resistance to radio-/chemotherapy.4,5 Moreover, GC is a heterogeneous cancer, and several genes and biological pathways alter the process of tumorigenesis. Therefore, it is necessary to construct a stable biological pathway-based prediction model to lead to diagnosis and prognosis prediction for clinical work.
Several studies have already reported the activation of diverse biological processes via experimental results or bioinformatic analysis. Yeon et al6 reported that the miR-181b-5p/CAGE/S1PR1 axis can alter autophagic flux, resulting in enhanced sensitivity to anticancer drugs. Wang et al7 revealed that cisplatin resistance was transmitted by the exosomal lncRNA HOTTIP, which targets the miR-218/HMGA1 axis in cisplatin-sensitive cells. Other studies also illustrated a prognostic model for GC patients. Wang et al8 revealed that early B cell factors are prognostic biomarkers for GC patients, and 5-fluorouracil, methotrexate, and vorinostat are suitable to inhibit the function of early B cell factors and suppress the process of tumorigenesis. Li et al9 developed a prognostic model, IDOScore, based on the expression levels of TAP2, SERPINB5, LTBP1 and LAMC1. IDOScore inversely reflects the response to immunotherapy in GC patients and acts as a harmful prognostic factor.
Recently, most prognostic models have only focused on the levels of several genes, miRNAs, lncRNAs, gene mutations, or DNA methylation; however, the activation status of biological pathways is more stable and can reflect the comprehensive inner condition of tumours. In the current study, we first identified the most highly activated pathways in two datasets of GC patients and further constructed a prognostic model based on the hub genes in the pathways to establish a stable prognostic model for GC patients.
Methods
Summary of Patients
We collected samples from the Cancer Genome Atlas Stomach Adenocarcinoma cohort (TCGA-STAD cohort, https://www.cancer.gov) and GSE62254 cohort (Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/). The mRNA expression of the TCGA-STAD cohort was downloaded from the Genomic Data Commons (GDC) via the “TCGAbiolinks” package. The clinical information, especially the overall survival (OS) time and status, was also obtained through the “TCGAbiolinks” package. For the GSE62254 cohort, we downloaded the original expression data and annotated the gene symbols via the corresponding GPL570 platform. After the filtration of patients with the available data for both gene expression and clinical information, a total of 294 GC patients from the TCGA-STAD cohort and 300 GC patients from GSE62254 were enrolled for subsequent analysis. The immunohistochemistry slide was obtained from the Human Protein Atlas (https://www.proteinatlas.org/).
GSEA
Gene set enrichment analysis (GSEA) is a computational method that determines whether a set of genes shows statistically significant differences between two groups. We employed GSEA to first compare the diverse activated signalling pathways between dead GC patients and living patients in the TCGA-STAD cohort and GSE62254 cohort. The background file of molecular signature gene sets was downloaded from MSigDB, C2, and KEGG. Then, the pathways that were both activated or inactivated in the two cohorts were considered for subsequent calculation.
Removal of Batch Effects
When obtaining the gene expression profile, batch effects will occur due to differences in nonbiological factors. To make the gene expression profiles of the two cohorts more similar, we used the ComBat method described in the “sva” package to remove the batch effects between these two cohorts.
Identification of the GC Prognostic Index
A total of 218 genes from the ECM receptor interaction pathway and focal adhesion pathway were extracted for the identification of the GC prognostic index (GCPI). The least absolute shrinkage and selection operator (LASSO) regression analysis was subsequently performed by the “glmnet” package to generate a statistical model with high prediction accuracy and interpretability. The LASSO analysis selected genes, and corresponding coefficients were used to calculate the risk score via the following formula:
where 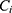 is the coefficient of the gene and
is the coefficient of the gene and 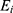 is the expression value of the corresponding gene.
is the expression value of the corresponding gene.
Search for Potential Therapeutic Strategies
We obtained the gene expression profile of 47 melanoma patients who received anti-cytotoxic T lymphocyte-associated protein 4 (anti-CTLA4) or anti-programmed death-1 (anti-PD1) and collected the response information. Meanwhile, the gene expression profile and clinical response information from another cohort were also recorded, comprising 248 bladder cancer patients.10–13 Submap analysis via the GenePattern platform was applied to compare the gene expression distribution between the defined subgroups in the current study and the responders to immunotherapy to reveal the patients who might respond to immunotherapy.
Statistics
All statistical analyses were conducted by R (Version: 4.0.2) using a two-sample Mann–Whitney test for continuous data, which is represented as the mean ± standard deviation (SD), and Fisher’s exact test for categorical data. A two-tailed P value less than 0.05 was considered statistically significant. For survival analyses, a Kaplan–Meier curve was generated for survival rates of patients with difference detection by the Log rank test. Cox regression was used to calculate the hazard ratios (HRs) and 95% confidence intervals (CIs) for OS. In addition, the prediction efficiency of GCPI for OS was examined using ROC analyses with the R package “pROC”.14 The forest displayed the prognostic value of GCPI, and other features were assessed by the function ggforest in the R package “surviminer”. The heatmap displayed the selected genes and clinical features performed by the R package “pheatmap”. We reproduced the risk subtypes with the minimal gene set in the training TCGA-STAD cohort by the random forest (RF) algorithm via “varSelRF” R package. The correlation between two continuous data points was evaluated by Pearson correlation analysis.
Results
Activation of Extracellular Matrix-Associated Pathways in GC Patients
We first revealed the activated signalling pathways in dead GC patients compared with living GC patients in both the GSE62254 and TCGA-STAD datasets. With the GSEA and a nominal P value less than 0.05, we obtained 24 pathways that were significantly activated in the GSE62254 dataset (Figure 1A), as well as 21 pathways in the TCGA-STAD dataset (Figure 1B). The detailed count number, normalized enrichment score (NES) and nominal P value are displayed in Table 1. After merging the coactivated pathways in both the GSE62254 and TCGA-STAD cohorts, we selected the KEGG Focal Adhesion and KEGG ECM Receptor Interaction pathways for subsequent analysis (Figure 1C). The detailed display of three key pathways in these two cohorts is given in Figure 1D and E.
 |
Table 1 Activated Pathways in Patients That Died from Gastric Cancer |
Construction of the Prognostic Model for GC Patients
We obtained the gene symbols of Focal Adhesion and ECM Receptor Interaction pathways from KEGG. After removing the duplicated genes, a total of 218 genes were enrolled for the LASSO analysis. At the beginning of the analysis, we removed the batch effect to make the selection of genes available in both cohorts. As shown via the principal component analysis (PCA) plot, we observed gaps between the GSE62254 and TCGA-STAD cohorts before removal (Figure 2A) and after removal (Figure 2B). The minimum lambda value of 0.02076 was defined as the cut-off point to minimize the mean cross-validated error (Figure 2C), and a total of 46 genes were identified (Figure 2D). The expression distribution of the 46 genes and clinical features in both the GSE62254 and TCGA-STAD cohorts are illustrated in Figure 2E. The 46 genes automatically separated into two groups, oncogenes and tumour suppressors. Regarding the clinical features, it was also clear that more dead patients in the high-risk group accounted for 54.5% (P < 0.001), and the average OS time for low-risk patients was 40.00 ± 34.44 months but sharply decreased to only 23.52 ± 27.01 months among patients with high-risk scores (P < 0.001). The distribution of sex and age showed no difference between these two subgroups, while more patients with advanced tumour stage belonged to the high-risk group, such as Stage IV (23.2% vs 13.1, P value for Chi square test: 0.004, Table 2).
 |
Table 2 Summary of the Clinical Features in Low-Risk and High-Risk Groups |
The High-Risk Group Demonstrated a Significantly Lower OS Time
To evaluate the prognostic value of these 46 genes and the LASSO analysis integrated risk score, we employed the K-M plot, Cox regression test and ROC curve. The prognostic value of each single gene is shown in Figure S1. The integrative risk score had wonderful prognostic value, and patients were separated into high-risk and low-risk groups based on the median value of the risk score. Then, we observed that patients in the high-risk group faced a significantly poorer prognosis than those in the low-risk group (P < 0.001, HR: 4.62, 95% CI: 3.447–6.183, Figure 3A), and the AUC value reflected the moderate prognostic value of the score (AUC: 0.694, 95% CI: 0.651–0.737, Figure 3B). We also evaluated the prognostic value in different clinical subgroups. It is promising that the risk score can reveal the high-risk patients among all patients in any of the clinical subgroups, including age ≤ 65 (P < 0.001), age > 65 (P < 0.001), male sex (P < 0.001), female sex (P < 0.001), and stage I (P = 0.002)/II (P < 0.001)/III (P < 0.001)/IV (P < 0.001, Figure S2). Furthermore, we evaluated the prognostic value of the risk score in the TCGA-STAD and GSE62254 cohorts. Patients in the high-risk group of the TCGA-STAD cohort had an approximately 3-fold higher risk of death than the low-risk group, with a 95% CI of 1.794 to 5.017 (P < 0.001, Figure 3C), and an AUC value of 0.672, with a 95% CI of 0.600 to 0.744 (Figure 3D). For the GSE62254 cohort, the prognostic function was more effective, the HR value was 4.94 (95% CI: 3.413–7.165, Figure 3E), and the AUC value was as high as 0.834 (95% CI: 0.789–0.879, Figure 3F).
Risk Score is an Independent Prognostic Factor for GC Prognosis
To further investigate whether the prognostic value of the risk score might be impacted by other clinical features, we performed multi-Cox regression analysis and proposed the combined nomogram of risk score and clinical features to elevate the performance. For the TCGA-STAD cohort, risk score (P < 0.001, HR: 2.77, 95% CI: 1.61–4.75), tumour stage IV (P = 0.002, HR: 3.91, 95% CI: 1.63–9.45), and age (P = 0.007, HR: 1.04, 95% CI: 1.01–1.07) were independent prognostic factors, as obtained from the results of multi-Cox regression analysis (Figure 4A). After combining the clinical features, the prognostic value of the single risk score increased from 0.657 to 0.741 (Figure 4B). Similar results were also observed in the GSE62254 cohort. The independent prognostic factors included age (P = 0.006, HR: 1.02, 95% CI: 1.01–1.04), stage III (P = 0.015, HR: 3.16, 95% CI: 1.25–8.02), stage IV (P < 0.001, HR: 6.98, 95% CI: 2.76–17.63) and risk score (P < 0.001, HR: 4.34, 95% CI: 2.91–6.49) (Figure 4C). The combined nomogram elevated the prognostic AUC value from 0.824 to 0.862 (Figure 4D).
Increased Risk Score Positively Associated with the Infiltration of Dendritic Cells
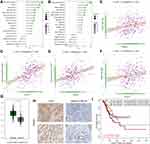
Several studies have already reported the association between poor prognosis of GC patients and immune infiltration. We calculated the correlation between the risk score and infiltration of 28 immunocytes in both the TCGA-STAD (Figure 5A) and GSE62254 (Figure 5B) cohorts. Most of the immunocytes were positively associated with an increased risk score, and there is no doubt that macrophages were included. We focused on the infiltration of dendritic cells, which displayed the highest correlation in both cohorts (TCGA-STAD: P < 0.001, R = 0.49, Figure 5C; GSE62254: P < 0.001, R = 0.39, Figure 5D). We also revealed that the activation of extracellular matrix pathways was positively associated with the infiltration of dendritic cells (Figure S3). In the tumour microenvironment, IFN-α is mostly secreted by dendritic cells, and the IFNAR1 and IFNAR2 genes encode the protein subunit of the receptor of IFN-α.15,16 Therefore, we also assessed the association of IFNAR1 and IFNAR2 with the risk score in the TCGA-STAD cohort and obtained positive results (IFNAR2: P < 0.001, R = 0.25, Figure 5E; IFNAR1: P < 0.001, R = 0.21, Figure 5F), which means that the 46 tumour genes promoted the tumorigenesis of GC via the infiltration of dendritic cells and the IFN-α axis. Moreover, we observed that both the mRNA level (Figure 5G) and protein level (Figure 5H) of IFNAR1 increased in GC tumour tissues compared with normal tissues, indicating the participation of IFNAR1 in GC tumorigenesis. We also found that patients with a high level of IFNAR1 and high infiltration of dendritic cells had the worst clinical outcome (Figure 5I).
GC Patients with High-Risk Scores are More Effective at Anti-CTLA4 Therapy
Subsequently, we performed Submap analysis to reveal the response to potential immunotherapy in GC patient subgroups, which is widely used in several published studies. The gene expression profiles of 47 melanoma patients and 248 bladder cancer patients were downloaded as the background comparison, and then we compared the expression of key component genes in the GC patient subgroups with the gene expression of responders and nonresponders. In the TCGA-STAD cohort, we found that compared with melanoma patients, melanoma patients were more likely to respond to CTLA4 treatment but not PD1 treatment (Figure 6A) and showed no response to PD-L1 treatment (Figure 6B). Submap analysis in the GSE62254 cohort showed similar results; GC patients seemed to benefit more from anti-CTLA4 therapy but not PD1 or PD-L1 therapy (Figure 6C and D).
Newly Defined Signature Can Identify the High-Risk Patients in Proposed GC Molecular Subtypes
We compared the high- and low-risk groups with the pre-reported four GC molecular subtypes generated by the Cancer Genome Atlas Research Network. We observed that low-risk group contained more patients belonged to EBV and MSI group, while high-risk group contained more CIN and GS patients (P < 0.001, Figure 7A and B). After reviewed the prognosis results of four TCGA molecular types, we found that the EBV, CIN, MSI and GS types did not reveal the diverse prognosis of GC patients. But the current study provides new sight for the prognosis of different molecular types. We found that although patients in the low-risk group contained less CIN and GS molecular subtype, but these two subtypes’ patients met the poor prognosis (Figure 7C). As to high-risk group, patients with the MSI features met the poor prognosis (Figure 7C). From the cross classification of risk score and TCGA-STAD molecular subtypes, we provided the new sight that the risk score can help to identify the patients with terrible prognosis in the inner of each molecular subtype.
We also used the RF method to select the minimal number of genes to reflect the diverse risk groups generated in our study. Finally, a total of 11 genes remained, including EGF, FLT1, GP9, ITGA2B, LAMC1, PAK6, SDC2, SHC4, TNN, VEGFB, VWF. We revealed the 11 genes in the TCGA-STAD training cohort, and further validated in the GSE62254 cohort, with an AUC value of 0.846, which reflected the stable value to identify patients with high- or low-risk (Figure 7D–F).
Discussion
The advanced tumour stage at first diagnosis and high resistance rate to radio-/chemotherapy result in a huge health burden of GC around the world. In the current study, we aimed to construct a stable prognostic model to identify GC patients with a high-risk of cancer-specific death to guide early intervention and precision treatment for GC patients.
In the GSE62254 and TCGA-STAD cohorts, we revealed that extracellular matrix-associated pathways were significantly activated in dead GC patients. The genes encoding ECM proteins and ECM-associated proteins are collectively known as the matrisome, which encompasses two main groups of genes: the core matrisome genes encoding collagens, glycoproteins, and proteoglycans, or matrisome-associated genes encoding ECM-affiliated proteins, ECM regulators, and secreted factors that are involved in the regulation or modulation of ECM functions.17,18 Several studies have already reported the function of the extracellular matrix in GC carcinogenesis. Han et al19 revealed that the inhibitory function of sulforaphane is achieved by boosting the maturation of miR-29a-3p to further downregulate two extracellular matrix components, COL3A1 and COL5A1. Liu et al20 found that high or low expression of ECM-related genes revealed different levels of tumour aggressiveness, and the zinc finger protein CTCF regulates ECM-related genes, thereby promoting GC cell growth and migration. Wei et al21 found that HOTAIR and COL5A1 were overexpressed in GC compared to normal controls, and HOTAIR regulates GC growth by sponging miR-1277-5p and upregulating COL5A1 via effects on the tumour microenvironment.
After the selection of LASSO analysis, a total of 46 genes were revealed to establish the prognostic models, such as PIK3CA, PIK3CB, ITGA11, and TNN. The PIK3CA and PIK3CB genes encode phosphatidylinositol-3-kinase (PI3K), and the PI3K axis has been shown to be activated in approximately one-third of human cancers. One of the malignancies that is most affected by this axis is GC. Zhao et al22 found that miR-522 can accelerate the progression of GC by targeting FOXO1 and regulating PI3K/AKT signalling. Among the different mechanisms that regulate EMT, IGF-1 signalling is the main regulator of this process in GC cells. Li et al23 reported that IGF-1 activation could increase the expression of ZEB2, and abrogation of the PI3K/AKT axis could downregulate ZEB2, resulting in the suppression of IGF-1-mediated EMT. Lu et al24 also found that abrogation of the PI3K/Akt axis could attenuate the oncogenic effect of miR-19a and thereby prevent the incidence of EMT in GC. As for ITGA11, Meng et al25 revealed its function in prostate cancer. ITGA11 protein levels were positively associated with advanced-stage disease, and silencing ITGA11 expression decreased cell migration and invasion.
The prognostic model was successfully established and validated in the TCGA-STAD and GSE62254 cohorts, especially in the GSE62254 cohort, and we obtained an AUC value as high as 0.834. Recently, several studies have reported prognostic models for GC patients based on gene expression profiles. Sun et al26 constructed a genomic instability-associated six-lncRNA signature with an integral AUC value of 0.681 that lacked external validation. Chen et al27 reported a ten-immune-related gene signature with a 1-year AUC of 0.681 and even failed to reach 0.57 in the GSE15459 cohort and 0.559 in the GSE84437 cohort; these results are not acceptable. Zhang et al28 reported an immune signature in the GSE62254 cohort, with an AUC value of only 0.793 in the entire cohort. Izumi et al29 constructed a gene expression-based 15-gene signature to distinguish lymph node status and obtained an AUC value of only 0.829 in the GSE62254 cohort. Li et al30 constructed a six gene-based prognostic model in the TCGA-STAD cohort; however, the AUC value they validated in GSE62254 was only 0.60–0.63. In addition, the prognostic model we established was an independent prognostic factor after adjusting for the clinical features of age, sex, stage, grade, and EBV status.
Immunotherapy is already applied in the clinical treatment of GC patients; therefore, it is important to select the appropriate patient to receive the therapy. In the KEYNOTE-059 trial, only 11.6% of recruited patients with advanced gastric/gastroesophageal junction cancer receiving pembrolizumab monotherapy were responders,31 and the objective response rate (ORR) of nivolumab in the ATTRACTION-2 trial was 11.2% for Asian patients.32 Zhou et al introduced a novel immunogenomic classification of the GC microenvironment and reported that patients with the immune activation subtype were ideal candidates to receive anti-PD-(L)1 immunotherapy, and those with the immunosuppressive subtype may be responders to combination therapy with anti-TGF-β antibodies. In the current study, we first compared the association of the risk score and 28 immunocytes and observed the highest association of dendritic cells. Further research revealed that the IFNAR1 protein was highly expressed in GC, and patients with high expression of IFNAR1 and infiltration of dendritic cells had the worst prognosis. In addition, we revealed that patients with a high-risk score were more suitable for anti-CTLA4 immunotherapy but not anti-PD1 or anti-PDL1 therapy.
This is the first study to generate a prognostic model with an activated signalling pathway for GC patients. We revealed that ECM-associated pathways were significantly activated in the two GC cohorts and then constructed a prognostic model similar to reeling silk from cocoons. Another advantage is that the newly defined risk score can act as the filter for patients who are more suitable for anti-CTLA4 immunotherapy. In contrast, several limitations should also be pointed out. First, these new findings need further validation in real-world patients, especially patients who receive immunotherapy. Second, biological experiments to confirm the function of dendrites and IFNAR1 are also necessary. We hope the findings in the current study can provide new insight for clinical treatment and basic research on GC patients.
Conclusion
We established and verified an extracellular matrix prognostic model of gastric patients. The model can be used to evaluate the risk of death of GC patients, as well as the response to anti-CTLA4 immunotherapy.
Data Sharing Statement
All data used in this work can be acquired from the GDC portal (https://portal.gdc.cancer.gov/), Gene-Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/).
Ethical Approval and Consent to Participate
Extra informed consent is not essential as the data were all obtained from a public database.
Acknowledgments
This work was supported by the Natural Science Foundation of Anhui Province,1808085MH247.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors have declared no conflicts of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
3. Thrift AP, El-Serag HB. Burden of gastric cancer. Clin Gastroenterol Hepatol. 2020;18(3):534–542. doi:10.1016/j.cgh.2019.07.045
4. Zheng P, Chen L, Yuan X, et al. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res. 2017;36(1):53. doi:10.1186/s13046-017-0528-y
5. Wei L, Sun J, Zhang N, et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol Cancer. 2020;19(1):62. doi:10.1186/s12943-020-01185-7
6. Yeon M, Kim Y, Pathak D, et al. The CAGE-MiR-181b-5p-S1PR1 axis regulates anticancer drug resistance and autophagy in gastric cancer cells. Front Cell Dev Biol. 2021;9:666387. doi:10.3389/fcell.2021.666387
7. Wang J, Lv B, Su Y, Wang X, Bu J, Yao L. Exosome-mediated transfer of lncRNA HOTTIP promotes cisplatin resistance in gastric cancer cells by regulating HMGA1/miR-218 axis. Onco Targets Ther. 2019;12:11325–11338. doi:10.2147/OTT.S231846
8. Wang Q, Liang J, Hu X, Gu S, Xu Q, Yan J. Early B cell factors involve in the tumorigenesis and predict the overall survival of gastric cancer. Biosci Rep. 2021;41(7). doi:10.1042/BSR20210055
9. Li L, Wang X. Identification of gastric cancer subtypes based on pathway clustering. NPJ Precis Oncol. 2021;5(1):1–17. doi:10.1038/s41698-021-00186-z
10. Lu X, Jiang L, Zhang L, et al. Immune signature-based subtypes of cervical squamous cell carcinoma tightly associated with human papillomavirus type 16 expression, molecular features, and clinical outcome. Neoplasia. 2019;21(6):591–601. doi:10.1016/j.neo.2019.04.003
11. Necchi A, Joseph RW, Loriot Y, et al. Atezolizumab in platinum-treated locally advanced or metastatic urothelial carcinoma: post-progression outcomes from the Phase II IMvigor210 study. Ann Oncol. 2017;28(12):3044–3050. doi:10.1093/annonc/mdx518
12. Meng J, Zhou Y, Lu X, et al. Immune response drives outcomes in prostate cancer: implications for immunotherapy. Mol Oncol. 2020. doi:10.1002/1878-0261.12887
13. Roh W, Chen PL, Reuben A, et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci Transl Med. 2017;9(379):eaah3560. doi:10.1126/scitranslmed.aah3560
14. Heagerty PJ, Lumley T, Pepe MS. Time‐dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344. doi:10.1111/j.0006-341X.2000.00337.x
15. Liao AP, Salajegheh M, Morehouse C, et al. Human plasmacytoid dendritic cell accumulation amplifies their type 1 interferon production. Clin Immunol. 2010;136(1):130–138. doi:10.1016/j.clim.2010.02.014
16. Wimmers F, Subedi N, van Buuringen N, et al. Single-cell analysis reveals that stochasticity and paracrine signaling control interferon-alpha production by plasmacytoid dendritic cells. Nat Commun. 2018;9(1):3317. doi:10.1038/s41467-018-05784-3
17. Socovich AM, Naba A. The cancer matrisome: from comprehensive characterization to biomarker discovery. Semin Cell Dev Biol. 2019;89:157–166. doi:10.1016/j.semcdb.2018.06.005
18. Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, Hynes RO. The extracellular matrix: tools and insights for the “omics” era. Matrix Biol. 2016;49:10–24. doi:10.1016/j.matbio.2015.06.003
19. Han S, Wang Z, Liu J, Wang HD, Yuan Q. miR-29a-3p-dependent COL3A1 and COL5A1 expression reduction assists sulforaphane to inhibit gastric cancer progression. Biochem Pharmacol. 2021;188:114539. doi:10.1016/j.bcp.2021.114539
20. Liu C, Deng L, Lin J, et al. Zinc finger protein CTCF regulates extracellular matrix (ECM)-related gene expression associated with the Wnt signaling pathway in gastric cancer. Front Oncol. 2020;10:625633. doi:10.3389/fonc.2020.625633
21. Wei Z, Chen L, Meng L, Han W, Huang L, Xu A. LncRNA HOTAIR promotes the growth and metastasis of gastric cancer by sponging miR-1277-5p and upregulating COL5A1. Gastric Cancer. 2020;23(6):1018–1032. doi:10.1007/s10120-020-01091-3
22. Zhao Y, Zhang J, Yang W, Yang Z, Zhou K. MicroRNA-552 accelerates the progression of gastric cancer by targeting FOXO1 and regulating PI3K/AKT pathway. J Oncol. 2021;2021:9966744. doi:10.1155/2021/9966744
23. Li H, Xu L, Zhao L, et al. Insulin-like growth factor-I induces epithelial to mesenchymal transition via GSK-3β and ZEB2 in the BGC-823 gastric cancer cell line. Oncol Lett. 2015;9(1):143–148. doi:10.3892/ol.2014.2687
24. Lu WD, Zuo Y, Xu Z, Zhang M. MiR-19a promotes epithelial-mesenchymal transition through PI3K/AKT pathway in gastric cancer. World J Gastroenterol. 2015;21(15):4564–4573. doi:10.3748/wjg.v21.i15.4564
25. Meng J, Guan Y, Wang Bet al. Molecular subtyping and prognostic assessment of prostate cancer based on consensus genes. Research Square. 2021. doi:10.21203/rs.3.rs-413181/v1
26. Sun J, Jiang Q, Chen H, et al. Genomic instability-associated lncRNA signature predicts prognosis and distinct immune landscape in gastric cancer. Ann Transl Med. 2021;9(16):1326. doi:10.21037/atm-21-3569
27. Chen T, Yang C, Dou R, Xiong B. Identification of a novel 10 immune-related genes signature as a prognostic biomarker panel for gastric cancer. Cancer Med. 2021;10(18):6546–6560. doi:10.1002/cam4.4180
28. Zhang C, Li D, Yu R, et al. Immune landscape of gastric carcinoma tumor microenvironment identifies a peritoneal relapse relevant immune signature. Front Immunol. 2021;12:651033. doi:10.3389/fimmu.2021.651033
29. Izumi D, Gao F, Toden S, et al. A genomewide transcriptomic approach identifies a novel gene expression signature for the detection of lymph node metastasis in patients with early stage gastric cancer. EBioMedicine. 2019;41:268–275. doi:10.1016/j.ebiom.2019.01.057
30. Li J, Pu K, Li C, Wang Y, Zhou Y. A novel six-gene-based prognostic model predicts survival and clinical risk score for gastric cancer. Front Genet. 2021;12:615834. doi:10.3389/fgene.2021.615834
31. Fuchs CS, Doi T, Jang RW, et al. Safety and efficacy of pembrolizumab monotherapy in patients with previously treated advanced gastric and gastroesophageal junction cancer: phase 2 clinical KEYNOTE-059 trial. JAMA Oncol. 2018;4(5):e180013. doi:10.1001/jamaoncol.2018.0013
32. Kang YK, Boku N, Satoh T, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, Phase 3 trial. Lancet. 2017;390(10111):2461–2471. doi:10.1016/S0140-6736(17)31827-5
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.