Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 18
Expression Profiles of circRNAs and Identification of hsa_circ_0007608 and hsa_circ_0064656 as Potential Biomarkers for COPD-PH Patients
Authors Yu J , Huang S, Shen W, Zhang Z , Ye S, Chen Y, Yang Y, Bian T, Wu Y
Received 26 July 2023
Accepted for publication 30 October 2023
Published 7 November 2023 Volume 2023:18 Pages 2457—2471
DOI https://doi.org/10.2147/COPD.S424712
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Richard Russell
Jinyan Yu,1,* Shulun Huang,1,* Weiyu Shen,1,* Zheming Zhang,1 Shugao Ye,2 Yuan Chen,2 Yue Yang,3 Tao Bian,3 Yan Wu3
1The Affiliated Wuxi People’s Hospital of Nanjing Medical University, Wuxi, Jiangsu, 214023, People’s Republic of China; 2Transplant Centre, the Affiliated Wuxi People’s Hospital of Nanjing Medical University, Wuxi, Jiangsu, 214023, People’s Republic of China; 3Department of Respiratory Medicine, the Affiliated Wuxi People’s Hospital of Nanjing Medical University, Wuxi, Jiangsu, 214023, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yan Wu; Tao Bian, Department of Respiratory Medicine, the Affiliated Wuxi People’s Hospital of Nanjing Medical University, Wuxi, Jiangsu, 214023, People’s Republic of China, Email [email protected]; [email protected]
Introduction: Pulmonary hypertension (PH) is a common complication of chronic obstructive pulmonary disease (COPD), which can worsen the prognosis and increase the mortality of COPD patients. Circular RNA (circRNA) has been discovered to participate in the occurrence and progression of PH in COPD and may have significant prospects for advanced diagnostics and prognosis evaluation. However, the expression profile of circRNAs in human lung tissues with definite diagnosis of COPD-PH remains to be further explored and validated.
Methods: Twelve human lung tissue samples (6 each from COPD-PH and control groups) were collected and subjected to high-throughput sequencing. QRT-PCR was performed to validate the differential expression levels of the top 10 dysregulated circRNAs in patients’ plasma samples, HPAECs and HPASMCs. Functional and pathway enrichment analysis on target genes was performed to explore the potential functions and pathways of those circRNAs. Hub genes obtained after conducting bioinformatics analysis on the predicted target mRNAs were verified by qRT-PCR in HPAECs and HPASMCs, and then we selected VCAN as a potential key gene involved in the pathogenesis of COPD-PH for immunohistochemistry validation in lung tissue.
Results: A total of 136 circRNAs (39 up-regulated and 97 down-regulated) were differentially expressed between the two groups. Following qRT-PCR validation, two circRNAs (hsa_circ_0007608 and hsa_circ_0064656) were believed to be involved in the pathogenesis. GO and KEGG pathway analysis suggested that these two DECs were mainly related to the celluar proliferation, migration and EndMT. PPI network revealed 11 pairs of key mRNAs. VCAM1, VCAN and THBS1, three hub mRNAs with the highest reliability among all, were validated and proven to be up-regulated in COPD-PH. We innovatively found that VCAN may be involved in COPD-PH.
Conclusion: This study identified the functional circRNAs, providing insights into the molecular mechanisms and predictions of COPD-PH, and may provide potential diagnostic biomarkers or therapeutic targets for COPD-PH.
Keywords: pulmonary hypertension, chronic obstructive pulmonary disease, circular RNA, RNA-sequencing, biomarker, bioinformatics analysis
Introduction
Chronic obstructive pulmonary disease (COPD) is a chronic inflammatory lung disease featured by airflow obstruction with high prevalence, morbidity, and mortality. It is reported that COPD is the third leading cause of death globally at present, and the incidence of COPD in adults over 40 years is more than 5%.1,2 Pulmonary hypertension (PH), a common complication of COPD, is characterized by aberrant pulmonary vasoconstriction and progressive pulmonary artery remodeling, which will finally result in right heart failure and even death.3,4 According to the statistics, the mortality rate of COPD patients with severe PH is around 12% per year, while the 5-year survival rate of patients with PH is less than 50%, reflecting the poor prognosis of COPD patients caused by PH.5 Moreover, COPD-PH is facing challenges in terms of diagnosis and treatment. Right heart catheterization (RHC), the gold standard for the diagnosis of PH, is rarely performed in clinical practice due to its invasion, while echocardiographic is not recommended for definite diagnosis in the standard for its high false positive rate.6 With efforts being made to develop drug therapies and interventional therapies, existing treatments can improve patients’ symptoms mainly by lessening the pulmonary vasoconstriction, but have no significant effects on reversing pulmonary vascular remodeling.7 Therefore, it is crucial that COPD-PH patients be diagnosed at an early stage and novel diagnostic biomarkers and molecular targets be explored to bring light into the mechanism of COPD-PH.
In recent years, researches have found that circular RNA (circRNA) participates in the initiation and development of PH. CircRNA is a type of single-stranded RNA molecule with covalently closed-loop structures. Compared to traditional linear RNA, circRNA is more stable, conserved and tissue-specific. Among all the functions, the best demonstrated function of circRNA is miRNA sponging. CircRNA can act as a sponge of the targeted miRNA and regulate its activity by repressing its expression, subsequently modulating the expression of their downstream target genes.8,9 Recent studies have shown that circRNA is closely related to the occurrence and development of respiratory diseases and have the potential to become novel diagnostic biomarkers as well as therapeutic targets for these diseases.10,11 Among them, our research group has identified that circSAV1 can play a key role in the pathogenesis of COPD.12 As well, there is evidence that circRNAs are also involved in the pathogenesis of COPD-PH. Zhou et al13 found that circHIPK3 promotes the proliferation of pulmonary arterial endothelial cells (HPAECs) and induces angiogenesis via the miR-328-3p/STAT3. Additionally, hsa_circNFXL1_009 was reported to attenuate hypoxia-induced proliferation, apoptotic resistance, and migration of and pulmonary arterial smooth muscle cells (HPASMCs).14 These researches indicate that circRNAs participate in the pathogenesis of COPD-PH by regulating the biological behaviour of pulmonary vascular cells. However, circRNA-related studies on COPD-PH patients are inadequate, on account of which, the underlying role of circRNA in COPD-PH has not been comprehensively explained.
Bioinformatics tools have been widely applied in oncology to identify genetic changes and new potential biomarkers associated with various diseases.15–17 Likewise, researchers have tried to apply bioinformatics analysis in the study of PH to elucidate the potential functions of circRNAs related to PH, mostly based on databases and animal experiments.18–28 However, few studies have utilized clinical samples diagnosed by RHC, and researches for COPD-PH are even fewer. To the best of our knowledge, the present study is the first to profile circRNA expression of lung tissues from COPD patients with mean pulmonary artery pressure (mPAP)≥20 mmHg measured by right heart catheterization (RHC). A total of 136 differentially expressed circRNAs were identified between the COPD-PH and control groups. Subsequently, we validated the most 10 dysregulated circRNAs in patients’ serum and hypoxia-induced vascular cells by quantitative real-time polymerase chain reaction (qRT-PCR). Functional enrichment analyses were carried out to investigate the main functional pathways involved in the development of COPD-PH. After the prediction of miRNAs and mRNAs for candidate circRNAs, a circRNA-miRNA-mRNA interaction network was constructed. Our study identified 2 pivotal circRNAs, which have not been reported, and 3 downstream mRNAs, of which VCAN was firstly found to be up-regulated in COPD-PH, thus providing new diagnostic biomarkers and therapeutic target and might further the understanding of molecular mechanism of COPD-PH.
Materials and Methods
Ethics
The study procedure was approved by the Medical Ethics Committee of the Affiliated Wuxi People’s Hospital of Nanjing Medical University. All patients included in this study have signed the corresponding informed consent.
Sample Collection
We selected a total of 12 patients (6 patients were with COPD-PH and 6 patients without PH nor COPD), the lung tissues of the normal control group were from the patients underwent lobectomy due to pulmonary nodules, while the lung tissues of the COPD-PH group were from the patients underwent lung transplantation surgery at Wuxi People’s Hospital Affiliated to Nanjing Medical University from January 2020 to December 2020. We also collected blood samples from COPD-PH patients (n = 12, COPD-PH) and patients without PH nor COPD (n = 12, control) for further validation. Blood samples were centrifugated at 4°C, 3000 rpm for 10 min, and the extracted supernatant was stored at −80°C. Clinicopathological characteristics of the whole subjects involved are listed in Table S1.
Patients with COPD were diagnosed by post-bronchodilator forced expiratory volume 1s/forced vital capacity (FEV1/FVC) ratio lower than 70% and without other lung diseases, such as bronchiectasis, diffuse interstitial lung disease, cystic fibrosis or lung cancer. We collected enrolled patients’ RHC data to screen out COPD-PH subjects and selected patients with the mean pulmonary artery pressure (mPAP) ≥20 mmHg as subjects.28 All patients were excluded from non-COPD-related PH such as congenital heart disease, chronic thromboembolic pulmonary hypertension (CTEPH) and arterial pulmonary hypertension (PAH).
Cell Culture
Human pulmonary arterial endothelial cells (HPAECs) were purchased from ScienCell Research Laboratory and cultured in endothelial culture medium. Pulmonary arterial smooth muscle cells (HPASMCs) were purchased from iCell Research Laboratory cultured in smooth muscle culture medium. Control group cells were cultured at 37°C in a humidified incubator with 5% CO2, while hypoxia group was incubated in a Tri-Gas Incubator (Heal Force, Shanghai, PR China) with a gas mixture containing 95% N2, 4% CO2, 1% O2 for 48h. Cells were trypsinized and passaged at 90% confluence, and the studies were performed on cells at passages 3–6.
RNA Isolation and Quality Control
Total RNA was isolated from samples using TRIzol (Invitrogen) according to the manufacturer’s protocols. The integrity and concentration of RNA samples were determined with Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA) and Nanodrop (Wilmington, DE, USA). RNA samples satisfied with the following parameters: OD260/280 between 1.8 and 2.1, OD260/280 >1.8, were subjected to the subsequent analysis.
Library Construction and circRNA Data Analysis
A total of 5μg RNA from samples were used as input material for the RNA sample preparations. The extracted RNA was treated with Epicentre Ribo-zero™ rRNA Removal Kit (Epicentre, USA) and RNase R (Epicentre, USA) to remove ribosomal and linear transcripts respectively. Sequencing libraries were generated using NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, USA) according to the manufacturer’s protocol. End products were purified by AMPure XP system, and library was assessed for quality and quantified on the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Qualified reads were mapped against human genome references using find_circ and CIRI.4,29 Subsequently, the circRNA expression profiles between the two groups were evaluated using fold-change filtering and t-test, with a statistical significance defined by fold-change value of ≥2.0 and a P-value of <0.05. The sequencing procedures and analyses were performed by Novogene Biotech (Beijing, China).
Quantitative Real-Time Polymerase Chain Reaction Validation (qRT-PCR) Validation
To further validate the RNA-Seq results, the top 10 DECs and three hub mRNAs were selected for qRT-PCR analysis. Two circRNA primers were not successfully designed, and they were removed in the following experiment. The specific primers of each circRNA were designed by Sangon Biotech (Shanghai, China). Primers are presented in Table 1. Total RNA was extracted as mentioned above, and the cDNA was synthesized from 1000ng of extracted RNA using PrimeScriptTMRT reagent Kit (Takara, Japan). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using TB GreenTM Premix Ex TaqII (Takara, Japan) on an ABIPRISM 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) with the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an internal control. The PCR conditions were 95°C for 30 min, followed by 40 cycles of 95°C for 5 s and 60°C for 34s. The 2−ΔΔt method was used to determine the relative quantification of gene expression levels. The qRT-qPCR for each target gene was repeatedly performed in triplicate.30
 |
Table 1 Primers of Validated circRNAs |
Prediction of Target miRNAs and mRNAs and Construction of circRNA-miRNA-mRNA Network
Previous studies have verified that circRNA contains miRNA response elements (MREs) and can function as miRNA sponges to regulate their potential functions through competing endogenous RNA (ceRNA). To further clarify the role of circRNAs in COPD-PH, we chose 10 circRNAs with the most obvious expression difference, including five up-regulated and five down-regulated circRNAs, and predicted the miRNA and mRNA targets of these circRNAs based on Miranda (www.miranda.org/) and TargetScan (www.targetscan.org/) application.31 Then, a circRNA-miRNA-mRNA network was constructed by Cytoscape software (version 3.2.0; https://cytoscape.org/).
Bioinformatics Analysis
The functions of the target genes were further analyzed by using Gene Ontology (http://www.geneontology.org) and were classified into three domains: biological process (BP), molecular function (MF) and cellular component (CC). GO terms with corrected P-value less than 0.05 were considered to be significantly enriched in differentially expressed genes. In addition, Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) was used to identify the biological pathways of target genes. Gene Ontology annotation and KEGG analysis were undertaken using an R package named clusterProfiler.32
Establishment of the Protein–Protein Interaction (PPI) Network
We acquired data of key mRNAs and constructed the PPI network via STRING database (https://string-db.org/). Then, we construct a PPI network using Cytoscape.33 Then, we used the Cytohuba, a Cytoscape plugin, to rank the 11 pairs of mRNAs according to degree and the correlation with HPH to screen out hub genes.
Immunohistochemistry
Immunohistochemical (IHC) staining was used to detect the expression of VCAN in lung vessels. In short, lung tissue was embedded in paraffin, and paraffin sections were prepared. The sections were dewaxed and dehydrated and microwaved in antigen repair solution for 20 minutes. Subsequently, BSA blockade was performed, and sections were incubated with primary antibodies to resist VCAN (1:200, ab177480) overnight at 4 °C. After washing with PBS, incubate with the second antibody at room temperature for one hour. All sections were observed and photographed using an optical microscope (Leica, Wetzlar, Germany).
Statistical Analysis
All of the statistical analyses were performed using GraphPad Prism v6.01 (GraphPad Software, CA, USA), R Studio version 3.2.1 (http://www.r-project.org/) and Microsoft Excel (Microsoft, DC, USA). The difference between 2 groups were assessed by Student’s s t-test. P-value <0.05 was considered as statistically significant.
Results
Analysis of circRNA Expression Profile in COPD-PH Patients
In our RNA sequence, a total of 11,193 circRNAs were identified in two groups of samples. All data were analyzed according to the workflow (Figure 1).
 |
Figure 1 The workflow of study. Abbreviations: GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein–protein interaction. |
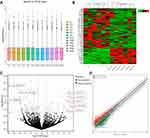
A box plot of the normalized expression of genes in all samples showed the credibility of our data (Figure 2A). A cluster heatmap was used to assess the expression variation of circRNAs between two groups (Figure 2B), in which the high and low expression of circRNAs were marked in red and green, respectively. A volcano plot (Figure 2C) and a scatter plot (Figure 2D) were constructed to visualize the differential expression of circRNAs between the two groups. In total, 136 DECs were identified between the COPD-PH and control groups, of which 39 circRNAs were up-regulated and 97 circRNAs were down-regulated. The top 10 DECs are listed in Table 2.
 |
Table 2 The Top 10 Differentially Expressed circRNAs with the Most Significant Difference in the COPD-PH |
qRT-PCR Validation of circRNAs Expression
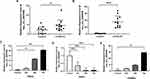
We selected the top 10 DECs based on the circRNA expression profiles for further validation in 12 pairs of subjects’ serum. Two circRNA primers were not successfully designed, and they were removed in the following experiment. The circRNAs involved in the validation included five up-regulated circRNAs (including hsa_circ_0007608, hsa_circ_0076410, hsa_circ_0011572, hsa_circ_0026227, hsa_circ_0064656) and three down-regulated circRNAs (including hsa_circ_0007121, hsa_circ_0019061, hsa_circ_0086483). Analysis by qRT-PCR confirmed three up-regulated circRNAs in serum, which were consistent with the results of the circRNA microarray. More specifically, it was found that hsa_circ_0007608, hsa_circ_0076410 and hsa_circ_0064656 were significantly up-regulated in COPD-PH group compared with that in control group (Figure 3A and B). Furthermore, we found that the expression of hsa_circ_0007608 was constantly up-regulated with the prolongation of hypoxia in HPAECs (Figure 3C), which is consistent with the results verified in serum. Interestingly, hsa_circ_0064656 was confirmed to express differently in different cells. When chronic hypoxia occurs in pulmonary vascular cells, hsa_circ_0064656 decreased significantly in HPAECs (Figure 3D), while increased in HPASMCs (Figure 3E). This phenomenon suggests that hsa_circ_0064656 may be involved in the intercellular signal transduction between HPAECs and HPASMCs in pulmonary vessels under hypoxia environment during COPD-PH pathogenesis.
Identification of Key mRNAs Based on Interaction of Predicted mRNAs and DEmRNAs
CircRNAs have been known to sponge miRNAs, leading to the derepression of miRNA targets and act as “miRNA sponges” within the cellular RNA network. We selected the two most dysregulated circRNAs and then mapped them with the miRanda database to obtain the targeted miRNAs of the two circRNAs. We screened out a total of 13 miRNAs and 4099 target mRNAs of miRNAs were identified via online databases.
As mentioned above, we also evaluated differentially expressed mRNAs (DEmRNAs) between the two groups. A cluster heatmap (Figure 4A) and a volcano plot (Figure 4B) were used to show the differential expression of mRNAs between the two groups. In order to find the key mRNAs involved in COPD-PH pathogenesis more accurately, we selected 35 mRNAs through the interaction analysis with target mRNAs and DEmRNAs (Figure 4C). Finally, the regulatory network of two circRNAs, 13 miRNAs and 35 mRNAs was constructed (Figure 4D).
GO and KEGG Analyses of circRNA-Targeting Genes
In order to accurately predict the key role of circRNA in the pathogenesis of COPD-PH, the linear transcripts of the corresponding genes of the target of circRNA were further evaluated through GO and KEGG pathway analysis. The GO analysis results from the Biological Process(BP), Cellular Component (CC) and Molecular Function (MF) domains (Figure 5). The top 10 dysregulated GO processes in each domain were analyzed and ranked according to p-value (p < 0.05). The top 3 enriched GO terms were regulation of intracellular signal transduction, regulation of phosphate metabolic process, regulation of phosphorus metabolic process in BP; extracellular matrix (ECM), interphotoreceptor matrix and extracellular region part in CC; calcium ion binding, metalloendopeptidase activity and ion binding in MF.
KEGG pathway enrichment analysis was also carried out to explore potential biological processes and pathways enriched by the target genes of circRNAs. The top 5 enriched KEGG pathways included NF-κB signaling pathway, malaria, TGF-β signaling pathway, MAPK signaling pathway and TNF signaling pathway (Figure 6).
 |
Figure 6 A dot plot of top 10 KEGG pathway of linear counterparts of DECs in COPD-PH. KEGG analysis according to rich factor, q-value and the number of enriched genes. |
Identification and Validation of Hub mRNAs
To determine the interactions between the downstream mRNAs of hsa_circ_0007608 and hsa_circ_0064656 and identify hub mRNAs in COPD-PH, we further made correlation analysis of 35 key mRNA (Figure 7A). Among them, 11 pairs of protein interactions were screened according to the reliability greater than 0.40 (Figure 7B). We continued to analyze the correlation between these key mRNAs and COPD-PH and finally screened three hub mRNAs with high reliability greater than 0.9 and closely related to COPD-PH. They included vascular cell adhesion molecule 1 (VCAM1), versican (VCAN), thrombospondin 1 (THBS1). We verified the three hub mRNAs in two kinds of pulmonary vascular cells by qRT-PCR and proved that the expression of these three hub mRNAs was indeed up-regulated in pulmonary vessel cells after 48 hours of hypoxia treatment (Figure 7C-H). Furthermore, we innovatively confirmed a significant upregulation of VCAN expression in the COPD-PH group of human pulmonary blood vessels by immunohistochemical analysis (Figure 7I and J).
Discussion
As a devastating cardiovascular complication of COPD, PH is inextricably linked with the increase of hospitalization rate, acute exacerbation and mortality of COPD patients.4 PH exhibits many cancer-like characteristics, including sustained cell proliferation and apoptosis resistance of pulmonary vascular cells such as pulmonary arterial endothelial cells, HPAECs and HPASMCs, dysregulated cellular metabolism and endothelial-to-mesenchymal transition (EndMT).29,30 However, the underlying mechanism of COPD-PH has not been clarified completely. With the rapid advances in gene sequencing technologies, several studies have identified some dysregulated circRNAs in PH, mostly based on animal experiments and public databases. However, there is still a lack of studies depending on clinical samples, and bioinformatics analysis on COPD-PH is rare. To achieve the high clinical value, we screened out the DECs from 12 human lung tissues. In this research, we identified 136 DECs between the COPD-PH and control groups, of which 39 circRNAs were up-regulated and 97 circRNAs were down-regulated, and the top ten dysregulated circRNAs were then validated by qRT-PCR in patients’ serum, HPAECs and HPASMCs. According to validation, hsa_circ_0007608 and hsa_circ_0064656 were selected for further construction of the circRNA-miRNA-mRNA regulatory network. Eleven key mRNAs were revealed by the PPI network and three of them (VCAM1, VCAN and THBS1) were validated in HPAECs and HPASMCs to be up-regulated in COPD-PH and functional enrichment analysis suggested that the hub genes were mainly involved in “intracellular signal transduction”, “extracellular matrix” and “calcium ion binding”. Taken together, the circRNA-miRNA-mRNA regulatory network that we constructed might help to elucidate the regulatory mechanisms of circRNA-related competing endogenous RNAs (ceRNA) in COPD-PH. Collectively, these results indicated that the ceRNA network constructed might help to elucidate the molecular mechanism of circRNAs in the pathogenesis of COPD-PH.
It is noteworthy that those two circRNAs have not been reported in any studies on PH so far, so there is an urgent need for more studies on the mechanisms of these 2 circRNAs in COPD-PH. Functional and pathway enrichment analysis was conducted to better understand their possible functions in COPD-PH. GO analysis manifested that hsa_circ_0007608 and hsa_circ_0064656 were chiefly enriched in the regulation of intracellular signal transduction, ECM and calcium ion binding. KEGG pathway results revealed that they were markedly enriched in NF-κB signaling pathway, malaria, TGF-β signaling pathway, MAPK signaling pathway and TNF signaling pathway. As shown in previous reports, the GO terms and KEGG pathways that hsa_circ_0007608 and hsa_circ_0064656 were enriched in were all related to the occurrence and regulation processes of PH. It is obvious that intracellular signal transduction is ubiquitous in the pathogenesis of COPD-PH.31,32 Existing studies also show that ECM is implicated in the development of pulmonary vascular remodeling and even precedes the onset of pulmonary hypertension.33 Moreover, critical regulators of ECM mainly including matrix metalloproteinases (MMPs) have been found to be closely related to the development of pulmonary vascular remodeling in PH.34 Additionally, release of calcium ions from the endoplasmic reticulum to the cytoplasm and mitochondria is an important mediator of cell proliferation, migration and mesenchymal transition, thereby affecting the contractility and resistance of pulmonary artery.35–37 As inflammation mediators, NF-κB, TGF-β, MAPK and TNF can regulate the functions of pulmonary vascular cells, including cellular proliferation, apoptosis and migration, contributing to pulmonary vascular remodeling.38–41 Interestingly, those 4 cytokines mentioned above also can induce or participate in the conversion process of endothelial cells into mesenchymal fibroblast-like cells, which is an important pathologic change of pulmonary vascular remodeling. More specifically, NF-κB, a cellular transcription factor regulating inflammation, plays a central role in inflammation-mediated EndMT, the stimulation of which and the subsequent production of cytokines are the key step in EndMT.42 TGF-β is the main inducer of EndMT, and the pathways through which TGF-β induced EndMT include MAPK.38,43 In addition, TNF, the most significant pro-inflammatory cytokines released during inflammation, is also related to the induction of EndMT.44 However, little is known about the correlation between malaria and the occurrence of COPD-PH, which needs further exploration. Combined with the above, the difference of hsa_circ_0064656 changing trends between the hypoxic HPAECs and HPASMCs could be attributed to the different roles these two cells play in EndMT under hypoxic condition. Taken together, we hypothesized that hsa_circ_0007608 and hsa_circ_0064656 might serve as key regulators in the pathogenesis of COPD-PH through these enriched pathways, especially through the regulation of EndMT, and possess the potential to become novel therapeutic targets, although the detailed biological mechanisms of those circRNAs and the interactions between the DECs and their target miRNAs need future in-depth studies.
Based on the prediction of downstream miRNAs and mRNAs, we constructed a circRNA-miRNA-mRNA regulatory network of the two validated DECs. Combined with the construction of PPI network and the qRT-PCR results, 3 hub genes including VCAM1, THBS1 and VCAN were recognized. According to previous reports, VCAM1, THBS1 contributes to the progression of pulmonary vascular remodeling. As a member of the immunoglobulin superfamily, VCAM1 contributes to endothelial dysfunction and is related to the inflammation of HPAECs.44,45 The study has also identified that THBS1 participates in the occurrence and development of PH.46,47 So far, there is no research to illustrate the role of VCAN in PH, indicating that VCAN might be a new discovery for PH and merits further exploration. Overall, the above results demonstrate that the ceRNA network is possibly involved in the pathogenesis of COPD-PH patients and may become novel diagnostic biomarkers and therapeutic targets for COPD-PH.
Previous studies have also explored the expression profile of circRNAs and the circRNA-miRNA-mRNA networks in PH. For instance, Wei et al21 identified 442 up-regulated DEGs and 84 down-regulated DECs from two microarray datasets (GSE53408 and GSE113439) and predicted 10 hub genes by PPI network. Additionally, 1216 DEGs in the blood of PH patients were screened out from four microarray datasets, and the functions of hub genes were demonstrated to be associated with multiple immune-related pathways in PAH by STRING-based investigation.20 Another study constructed the hypoxic pulmonary hypertension (HPH) rat model and found 15 significant DECs through High-throughput RNA sequencing.28 The qRT-PCR results revealed that 4 DECs (including circ_002723, circ_017779, circ_020592 and circ_020581) were consistent with the RNA-Seq data. Unlike previous researches, our study was based on clinical samples with definite diagnosis of COPD-PH. Besides, we have performed QRT-PCR in patients’ plasma samples, HPAECs and HPASMCs to validate the DECs and the hub genes in various aspects.
However, there are still a number of limitations in our study. Most of our results were based on computational prediction, which need further validation by molecular biological experiments. Besides, although cell experiments were performed to verify the reliability of the sequencing results, our study lacks in vivo experiments to further validate these results. Another limitation of the present study is the small sample size. Therefore, our findings await more comprehensive analyses and further in-depth studies to provide accurate evidence for prognostic and diagnostic biomarkers in COPD-PH.
Conclusion
To summarize, our study revealed the expression profile of circRNAs in COPD-PH patients, and established a circRNA-miRNA-mRNA interaction network. We discovered that hsa_circ_0007608 and hsa_circ_0064656 are the most notable markers of COPD-PH and may be regarded as potential targets for the treatment of COPD-PH. Moreover, VCAM1, THBS1 and VCAN are identified as the most reliable key mRNAs, of which VCAN might be a new discovery for the molecular mechanism of COPD-PH. These findings will provide bioinformatics basis for further research into the molecular mechanism of pulmonary hypertension in COPD patients and provide novel insight into the development of targeted drug therapy and diagnostic biomarkers.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Information File. The datasets generated or analysed during the current study are available in the Sequence Read Archive (SRA) database of NCBI [https://www.ncbi.nlm.nih.gov/sra/PRJNA913650].
Ethics Approval Statement
All donated lung tissue originates from patients underwent lung nodules or lung transplantation at Wuxi People’s Hospital Affiliated to Nanjing Medical University, and all experimental work was approved by the ethical review board of Wuxi People’s Hospital Affiliated to Nanjing Medical University. We confirm that lung donations were conducted with the full informed consent of the donor or their next-of-kin if deceased. This study was conducted in accordance with the tenets of the Declaration of Helsinki. The procedures were approved by the Ethics Committee of Nanjing Medical University (KY21033).
Author Contributions
All authors made a significant contribution to the work reported, whether in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all of these areas. All authors took part in drafting, revising or critically reviewing the article, gave final approval of the version to be published, have agreed on the journal to which the article has been submitted and agreed to be accountable for all aspects of the work.
Funding
This work was supported by the Natural Science Foundations of China (82173472), the Top Talent Support Program for young and middle-aged people of Wuxi Health Committee (BJ2020006), General Program of Wuxi Health Committee (M202032) and Wuxi Science and Technology Development Fund Project (Y20222007).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Ding X, Lin Q, Zhao J, et al. Synonymous mutations in TLR2 and TLR9 genes decrease COPD susceptibility in the Chinese Han population. Pulmonology. 2022;S2531-0437(22):220. doi:10.1016/j.pulmoe.2022.09.010
2. Ma C, Liao K, Wang J, et al. L-Arginine, as an essential amino acid, is a potential substitute for treating COPD via regulation of ROS/NLRP3/NF-κB signaling pathway. Cell Biosci. 2023;13:152. doi:10.1186/s13578-023-00994-9
3. Blanco I, Tura-Ceide O, Peinado VI, et al. Updated Perspectives on Pulmonary Hypertension in COPD. Int J Chron Obstruct Pulmon Dis. 2020;15:1315–1324. doi:10.2147/COPD.S211841
4. Vizza CD, Hoeper MM, Huscher D, et al. Pulmonary Hypertension in Patients With COPD: results From the Comparative, Prospective Registry of Newly Initiated Therapies for Pulmonary Hypertension (COMPERA). Chest. 2021;160(2):678–689.
5. Boucly A, Savale L, Jaïs X, et al. Association between Initial Treatment Strategy and Long-Term Survival in Pulmonary Arterial Hypertension. Am J Respir Crit Care Med. 2021;204(7):842–854. doi:10.1164/rccm.202009-3698OC
6. Yu X, Qin W, Lin X, et al. Synergizing the enhanced RIME with fuzzy K-nearest neighbor for diagnose of pulmonary hypertension. Comput Biol Med. 2023;165:107408. doi:10.1016/j.compbiomed.2023.107408
7. Zheng R, Xu T, Wang X, et al. Stem cell therapy in pulmonary hypertension: current practice and future opportunities. Eur Respir Rev. 2023;32(169):230112. doi:10.1183/16000617.0112-2023
8. Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691.
9. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
10. Luo H, Xiao T, Sun X, et al. The regulation of circRNA_kif26b on alveolar epithelial cell senescence via miR-346-3p is involved in microplastics-induced lung injuries. Sci Total Environ. 2023;882:163512. doi:10.1016/j.scitotenv.2023.163512
11. Chen Z, Zhu J, Zhou H, et al. The involvement of copper, circular RNAs, and inflammatory cytokines in chronic respiratory disease. Chemosphere. 2022;303(Pt 2):135005. doi:10.1016/j.chemosphere.2022.135005
12. Xia H, Wu Y, Zhao J, et al. N6-Methyladenosine-modified circSAV1 triggers ferroptosis in COPD through recruiting YTHDF1 to facilitate the translation of IREB2. Cell Death Differ. 2023;30(5):1293–1304. doi:10.1038/s41418-023-01138-9
13. Hong L, Ma X, Liu J, et al. Circular RNA-HIPK3 regulates human pulmonary artery endothelial cells function and vessel growth by regulating microRNA-328-3p/STAT3 axis. Pulm Circ. 2021;11(2):20458940211000234. doi:10.1177/20458940211000234
14. Jin X, Xu Y, Guo M, et al. hsa_circNFXL1_009 modulates apoptosis, proliferation, migration, and potassium channel activation in pulmonary hypertension. Mol Ther Nucleic Acids. 2020;23:1007–1019. doi:10.1016/j.omtn.2020.09.029
15. Yin L, Tang Y, Yuan Y. An Overview of the Advances in Research on the Molecular Function and Specific Role of Circular RNA in Cardiovascular Diseases. Biomed Res Int. 2022;2022:5154122. doi:10.1155/2022/5154122
16. Pan S, Chen Y, Yan Y, et al. The emerging roles and mechanisms of exosomal non-coding RNAs in the mutual regulation between adipose tissue and other related tissues in obesity and metabolic diseases. Front Endocrinol (Lausanne). 2022;13:975334. doi:10.3389/fendo.2022.975334
17. Zhang L, Wang C, Lu X, et al. Transcriptome sequencing of hepatocellular carcinoma uncovers multiple types of dysregulated ncRNAs. Front Oncol. 2022;12:927524. doi:10.3389/fonc.2022.927524
18. Zheng H, Hua J, Li H, et al. Comprehensive analysis of the expression of N6-methyladenosine RNA methylation regulators in pulmonary artery hypertension. Front Genet. 2022;13:974740. doi:10.3389/fgene.2022.974740
19. Han X, Li C, Yang P, et al. Potential mechanisms of Qili Qiangxin capsule to prevent pulmonary arterial hypertension based on network pharmacology analysis in a rat model. Ann Transl Med. 2022;10(8):453. doi:10.21037/atm-22-901
20. Wang J, Uddin MN, Wang R, et al. Comprehensive analysis and validation of novel immune and vascular remodeling related genes signature associated with drug interactions in pulmonary arterial hypertension. Front Genet. 2022;13:922213. doi:10.3389/fgene.2022.922213
21. Wei RQ, Zhang WM, Liang Z, et al. Identification of Signal Pathways and Hub Genes of Pulmonary Arterial Hypertension by Bioinformatic Analysis. Can Respir J. 2022;2022:1394088. doi:10.1155/2022/1394088
22. Wang RR, Yuan TY, Chen D, et al. Dan-Shen-Yin Granules Prevent Hypoxia-Induced Pulmonary Hypertension via STAT3/HIF-1α/VEGF and FAK/AKT Signaling Pathways. Front Pharmacol. 2022;13:844400. doi:10.3389/fphar.2022.844400
23. Tang S, Liu Y, Liu B. Integrated bioinformatics analysis reveals marker genes and immune infiltration for pulmonary arterial hypertension. Sci Rep. 2022;12(1):10154. doi:10.1038/s41598-022-14307-6
24. Tu J, Jin J, Chen X, et al. Altered Cellular Immunity and Differentially Expressed Immune-Related Genes in Patients With Systemic Sclerosis-Associated Pulmonary Arterial Hypertension. Front Immunol. 2022;13:868983. doi:10.3389/fimmu.2022.868983
25. Lin W, Tang Y, Zhang M, et al. Integrated Bioinformatic Analysis Reveals TXNRD1 as a Novel Biomarker and Potential Therapeutic Target in Idiopathic Pulmonary Arterial Hypertension. Front Med. 2022;9:894584. doi:10.3389/fmed.2022.894584
26. Wang S, Liu Y, Wang Q, et al. Utilizing Network Pharmacology and Molecular Docking Integrated Surface Plasmon Resonance Technology to Investigate the Potential Targets and Mechanisms of Tripterygium wilfordii against Pulmonary Artery Hypertension. Evid Based Complement Alternat Med. 2022;2022:9862733. doi:10.1155/2022/9862733
27. Wei R, Chen L, Li P, et al. IL-13 alleviates idiopathic pulmonary hypertension by inhibiting the proliferation of pulmonary artery smooth muscle cells and regulating macrophage infiltration. Am J Transl Res. 2022;14(7):4573–4590.
28. Xu SL, Deng YS, Liu J, et al. Regulation of circular RNAs act as ceRNA in a hypoxic pulmonary hypertension rat model. Genomics. 2021;113(1 Pt 1):11–19. doi:10.1016/j.ygeno.2020.11.021
29. Humbert M, Guignabert C, Bonnet S, et al. Pathology and pathobiology of pulmonary hypertension: state of the art and research perspectives. Eur Respir J. 2019;53(1):1801887. doi:10.1183/13993003.01887-2018
30. Woo KV, Shen IY, Weinheimer CJ, et al. Endothelial FGF signaling is protective in hypoxia-induced pulmonary hypertension. J Clin Invest. 2021;131(17):e141467. doi:10.1172/JCI141467
31. Gallardo-Vara E, Ntokou A, Dave JM, et al. Vascular pathobiology of pulmonary hypertension. J Heart Lung Transplant. 2023;42(5):544–552. doi:10.1016/j.healun.2022.12.012
32. Xu WJ, Wu Q, He WN, et al. Interleukin-6 and pulmonary hypertension: from physiopathology to therapy. Front Immunol. 2023;14:1181987. doi:10.3389/fimmu.2023.1181987
33. Kucherenko MM, Sang P, Yao J, et al. Elastin stabilization prevents impaired biomechanics in human pulmonary arteries and pulmonary hypertension in rats with left heart disease. Nat Commun. 2023;14(1):4416. doi:10.1038/s41467-023-39934-z
34. Chelladurai P, Seeger W, Pullamsetti SS. Matrix metalloproteinases and their inhibitors in pulmonary hypertension. Eur Respir J. 2012;40(3):766–782. doi:10.1183/09031936.00209911
35. Guo X, Meng Y, Wang Y, et al. Mice lacking 1,4,5-triphosphate inositol type III receptor demonstrate inhibition of hypoxic pulmonary hypertension. Biochem Biophys Res Commun. 2022;629:165–170. doi:10.1016/j.bbrc.2022.09.036
36. Gabani M, Liu J, Ait-Aissa K, et al. MiR-204 regulates type 1 IP3R to control vascular smooth muscle cell contractility and blood pressure. Cell Calcium. 2019;80:18–24. doi:10.1016/j.ceca.2019.03.006
37. Balistrieri A, Makino A, Yuan JX. Pathophysiology and pathogenic mechanisms of pulmonary hypertension: role of membrane receptors, ion channels, and Ca2+ signaling. Physiol Rev. 2023;103(3):1827–1897. doi:10.1152/physrev.00030.2021
38. Pérez L, Muñoz-Durango N, Riedel CA, et al. Endothelial-to-mesenchymal transition: cytokine-mediated pathways that determine endothelial fibrosis under inflammatory conditions. Cytokine Growth Factor Rev. 2017;33:41–54. doi:10.1016/j.cytogfr.2016.09.002
39. Silva GF, da Silva JS, de Alencar AKN, et al. Novel p38 Mitogen-Activated Protein Kinase Inhibitor Reverses Hypoxia-Induced Pulmonary Arterial Hypertension in Rats. Pharmaceuticals. 2022;15(7):900. doi:10.3390/ph15070900
40. Pugliese SC, Poth JM, Fini MA, et al. The role of inflammation in hypoxic pulmonary hypertension: from cellular mechanisms to clinical phenotypes. Am J Physiol Lung Cell Mol Physiol. 2015;308(3):L229–L252. doi:10.1152/ajplung.00238.2014
41. Maleszewska M, Moonen JR, Huijkman N, et al. IL-1β and TGFβ2 synergistically induce endothelial to mesenchymal transition in an NFκB-dependent manner. Immunobiology. 2013;218(4):443–454. doi:10.1016/j.imbio.2012.05.026
42. Chen PY, Qin L, Baeyens N, et al. Endothelial-to-mesenchymal transition drives atherosclerosis progression. J Clin Invest. 2015;125(12):4514–4528. doi:10.1172/JCI82719
43. Adjuto-Saccone M, Soubeyran P, Garcia J, et al. TNF-α induces endothelial-mesenchymal transition promoting stromal development of pancreatic adenocarcinoma. Cell Death Dis. 2021;12(7):649. doi:10.1038/s41419-021-03920-4
44. Jang AJ, Chang SS, Park C, et al. PPARγ increases HUWE1 to attenuate NF-κB/p65 and sickle cell disease with pulmonary hypertension. Blood Adv. 2021;5(2):399–413. doi:10.1182/bloodadvances.2020002754
45. Iranmehr A, Stobdan T, Zhou D, et al. Novel insight into the genetic basis of high-altitude pulmonary hypertension in Kyrgyz highlanders. Eur J Hum Genet. 2019;27(1):150–159. doi:10.1038/s41431-018-0270-8
46. Kumar R, Mickael C, Kassa B, et al. Interstitial macrophage-derived thrombospondin-1 contributes to hypoxia-induced pulmonary hypertension. Cardiovasc Res. 2020;116(12):2021–2030. doi:10.1093/cvr/cvz304
47. Moll M, Christmann RB, Zhang Y, et al. Patients with systemic sclerosis-associated pulmonary arterial hypertension express a genomic signature distinct from patients with interstitial lung disease. J Scleroderma Relat Disord. 2018;3(3):242–248. doi:10.1177/2397198318764780
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.