Back to Journals » Journal of Inflammation Research » Volume 14
Expression Pattern and Clinical Value of Key m6A RNA Modification Regulators in Abdominal Aortic Aneurysm
Authors Li T, Wang T, Jing J , Sun L
Received 30 June 2021
Accepted for publication 19 August 2021
Published 29 August 2021 Volume 2021:14 Pages 4245—4258
DOI https://doi.org/10.2147/JIR.S327152
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Tan Li,1 Tianlong Wang,2 Jingjing Jing,3,4 Liping Sun3,4
1Department of Cardiovascular Ultrasound, The First Hospital of China Medical University, Shenyang, People’s Republic of China; 2The First Clinical College of China Medical University, The First Hospital of China Medical University, Shenyang, People’s Republic of China; 3Tumor Etiology and Screening Department of Cancer Institute and General Surgery, The First Hospital of China Medical University, Shenyang, People’s Republic of China; 4Key Laboratory of Cancer Etiology and Prevention in Liaoning Education Department, The First Hospital of China Medical University, Shenyang, People’s Republic of China
Correspondence: Liping Sun
Tumor Etiology and Screening Department of Cancer Institute and General Surgery, The First Hospital of China Medical University, No. 155 NanjingBei Street, Heping District, Shenyang, Liaoning Province, 110001, People’s Republic of China
Tel +86-24-83282153
Fax +86-24-83282383
Email [email protected]
Background: Aberrant expression of N6-methyladenosine (m6A) RNA modification regulators plays a critical role in a variety of human diseases. However, their implication in abdominal aortic aneurysm (AAA) remains largely unknown. Herein, we sought to explore the general expression pattern and potential functions of m6A regulators in AAA.
Methods: We analyzed gene expression data of m6A regulators in human AAA and normal tissues from public GEO database. The R package and other tools such as m6A2Target database, Gene ontology (GO) functional and Kyoto encyclopedia of genes and genomes (KEGG) pathway analyses, gene set variation analysis (GSVA), Search Tool for the Retrieval of Interacting Genes (STRING), starBase, miRDB and Cytoscape software were applied for bioinformatics analysis to investigate the downstream molecular mechanisms and upstream regulatory mechanisms for distinctly expressed regulators. Quantitative real-time PCR (qRT-PCR) and enzyme-linked immunosorbent assay (ELISA) were performed to validate the expression of key m6A regulators in our collected human AAA specimens.
Results: We found that METTL14 and HNRNPC were the downregulated m6A regulators, and RBM15B was the upregulated methylation transferase in human AAA. The modified genes were primarily enriched in RNA catabolic process, regulation of translation, focal adhesion, transcription coregulator activity, ribosome, RNA transport, cell cycle, et al. METTL14, HNRNPC and RBM15B levels were correlated with the immune infiltration degree of Tcm, macrophages, mast cells, Tgd and NK CD56bright cells. A total of 154 and 76 target genes of three regulators were separately involved in body metabolism and autophagy in AAA disease, and their interactive relationships and hub genes were identified. The lncRNA-miRNA-mRNA interaction regulatory networks were also constructed for METTL14, HNRNPC and RBM15B. Based on our clinical tissue and serum samples, METTL14 exhibited lower expression levels in AAA and its rupture type, and low METTL14 expression was associated with high levels of WBC and CRP (all P < 0.05).
Conclusion: Our study presents an overview of the expression pattern and functional significance of m6A regulators in human AAA. Our findings will provide a valuable resource that may guide both mechanistic and therapeutic analyses about the role of key m6A regulators in AAA.
Keywords: abdominal aortic aneurysm, gene expression, m6A modification
Introduction
Abdominal aortic aneurysm (AAA) is a life threatening, complicated and multifactorial disease that develops because of the interaction between environmental risk factors and genetic predisposition.1 Inflammation, vascular smooth muscle cell (VSMC) apoptosis, extracellular matrix degradation, and increased oxidative stress are the major histopathological features of AAA.2 Immune inflammatory cells have been found to be in the central position of the whole process of AAA formation.3 Recent researchers suggest that body metabolism is closely related to the initiation and development of AAA, while autophagy is an essential process mediating proper vascular function.4,5 However, the complete mechanisms underlying AAA have not yet been elucidated.
As an emerging research field of post-transcriptional gene regulation, N6-methyladenosine (m6A) modification of RNA is widely present in eukaryotic cells, and has been focused for its important part in a variety of biological processes.6,7 M6A modification exhibits dynamic and reversible changes and is controlled by “writers” (methyltransferases), “erasers” (demethylases), and “readers” (methyl-binding proteins). M6A “writers” and “erasers” determine the m6A modification on a specific mRNA, and “readers” can recognize and bind to modification sites and transmit the code to downstream effectors.8 Many m6A regulators have been identified to play crucial roles in RNA methylation process, thereby affecting RNA functions, such as stability, translation, degradation, transport, and splicing.9 A number of studies showed that m6A regulators were implicated in the pathogenesis of various malignancies.10–12 Moreover, emerging evidence indicated that the dysregulation of m6A regulators was closely linked with the initiation and progression of specific cardiovascular diseases, including cardiac hypertrophy, ischemic heart disease, heart failure, vascular calcification, and pulmonary hypertension.13 Recently, He et al found a significant increase of m6A level in human AAA tissues and observed the aberrant expression of certain m6A regulators involved in m6A modification.14 Zhong et al revealed a vital role of METTL3/m6A-mediated miR-34a maturation in AAA formation.15 Although above findings may suggest a new view on the mechanism of m6A modification in AAA, the general expression characteristics of all m6A regulators in AAA and potential downstream/upstream molecular mechanisms for key m6A regulators are largely unclear.
Bioinformatics study on the data derived from gene chips has become a promising and efficient tool to screen the significant genetic or epigenetic variations associated with multiple diseases.16 For the first time, we attempted to use public dataset to evaluate the distinct expression pattern of m6A regulators in human AAA tissues. With further bioinformatics analyses, including differential expression, target gene prediction, functional enrichment analysis, protein–protein interaction (PPI) and competing endogenous RNA (ceRNA) network construction, we sought to construct the whole picture of m6A regulators in AAA disease. Finally, the expression and clinical significance of key AAA-related m6A regulators were validated in our collected samples. Our pilot study could provide clues for further researches concerning the molecular mechanism of epigenetic alterations in clinical AAA, which would be essential in identifying novel targets for early detection and treatment of AAA.
Materials and Methods
Data Collection and Screening of AAA-Related m6A Regulators
The mRNA expression profiling dataset GSE47472 (human), which included 14 AAA and 8 normal aortic samples, was downloaded from the GEO database. The differentially expressed genes (DEGs) were identified with the statistical threshold of |log2Fold Change (FC)|>0.7 and P<0.05 by the “limma” R package. Currently, seventeen genes, including ALKBH5, FTO, HNRNPA2B1, HNRNPC, IGF2BP1, IGF2BP2, IGF2BP3, METTL14, METTL3, RBM15, RBM15B, RBMX, YTHDC1, YTHDC2, YTHDF1, YTHDF2 and YTHDF3, are regarded as common m6A RNA methylation regulators. Then, DEGs and these m6A regulators were overlapped, and the overlapped genes were the differentially expressed m6A regulatory genes (DEMRGs) and defined as key AAA-related m6A regulators. Subsequently, a volcano plot was drawn to visualize the expression of DEGs and a heatmap diagram was constructed to exhibit the expression of m6A regulators in AAA and normal aortic tissues.
Functional Annotation of AAA-Related m6A Regulators
Based on GSE47472 dataset, Pearson correlation analysis was conducted with R to search the co-expressed genes of DEMRGs in AAA tissues using the cut-off criteria of |R|>0.5 and P<0.05. The m6A2Target database (http://m6a2target.canceromics.org) was applied to predict the target genes of DEMRGs.17 The genes obtained from the intersection of the co-expressed genes and predicted targets were identified as target genes modified by key AAA-related m6A regulators. Then, we performed Gene ontology (GO) enrichment analysis, which contained terms of biological processes (BP), cellular components (CC) and molecular functions (MF), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis using “clusterProfiler” within R package. The P. adjust and q value less than 0.05 were regarded as significant categories.
Because cell immunity was closely related with AAA risk, we used gene set variation analysis (GSVA) in the “limma” package in R to assess the score of each of the 24 types of immune cell subpopulations in AAA samples,18 and further analyzed the relationship between key m6A regulators and immune cells. Meanwhile, a total of 1660 human metabolic genes from the metabolic pathways were downloaded from KEGG database, while a total of 608 autophagy-related genes were integrated from HADb database and the GO_AUTOPHAGY gene set in GSEA website. Above genes were separately overlapped with target genes of AAA-related m6A regulators. Then, PPI network was analyzed on the overlapped genes using the Search Tool for the Retrieval of Interacting Genes (STRING) database with a confidence score >0.4 as the cutoff criterion, and subsequently visualized in Cytoscape software v3.8.1. Additionally, we used CentiScaPe plugin of Cytoscape to determine the hub genes associated with metabolism or autophagy in PPI.
CeRNA Network Construction for AAA-Related m6A Regulators
In the co-expression gene analysis based on GSE47472 dataset, we could obtain the long non-coding RNAs (lncRNAs) associated with DEMRGs. Then, the interactions between microRNAs (miRNAs) and DEMRGs were predicted using starBase and miRDB databases, and the overlapped miRNA-mRNA pairs were selected. In addition, miRNAs that targeted lncRNAs were predicted using starBase database. Finally, lncRNA-miRNA-mRNA networks were constructed based on the miRNAs interacting with both lncRNAs and DEMRGs, and the visualization of ceRNA networks was through Cytoscape software v3.8.1.
Our Clinical Specimens
Fourteen AAA tissue samples were obtained from full-thickness aneurysmal abdominal aortic tissues during the open surgical repair, whereas four non-aneurysmal abdominal aortas were acquired from organ donors during kidney transplantation in the First Hospital of China Medical University. All donated kidneys were from postmortem multiorgan donors whose living first-degree relatives voluntarily gave written informed consent, and this was conducted in accordance with the Declaration of Istanbul. After surgery, tissues were immediately separated into RNase-free centrifuge tubes and stored at −80°C until RNA isolation. In addition, the blood samples of 30 AAA patients and 30 gender and age matched controls were collected from our hospital with standardized sterile tubes, and centrifuged at 3500 r/min for 10 min at 4°C. The serum was separated and stored at −80°C until used further. The clinical parameters of AAA patients were collected from the medical records. We excluded the subjects who had Marfan syndrome, traumatic AAA, previous aortic surgery, malignant tumor, hematological disease, infectious disease, autoimmune disease, myocardial infarction or severe organ failure. The study was approved by the Ethics Committee of the First Hospital of China Medical University (Shenyang, China) and conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants.
Quantitative Real-Time PCR (qRT-PCR)
qRT-PCR was conducted to validate the DEMRGs in tissue samples. Under the manufacturer’s instructions, total RNA was reverse-transcribed to cDNA using PrimeScript™ RT reagent kit (Takara, Dalian, China) and qRT-PCR was performed using the SYBR Premix Ex Taq™ II (Takara, Dalian, China). β-actin was used as the reference gene and the relative expression levels of mRNAs in tissues were calculated with the 2−∆∆CT method. The specific primers are presented in Table 1.
 |
Table 1 Primer Sequences of qRT-PCR |
Enzyme-Linked Immunosorbent Assay (ELISA)
The protein expression of DEMRGs with differential mRNA levels was determined by ELISA in serum samples. And serum levels were detected using human ELISA Kits (Shanghai Enzyme Research Biotechnology Co., Ltd., China) strictly in line with the manufacturer’s instructions.
Statistical Analysis
The statistical analyses were carried out using SPSS 17.0 software. Quantitative data were expressed as means ± SD. The independent-sample t-test, Mann–Whitney U-test, χ2 test or Fisher’s exact test was applied for the comparisons between groups as appropriate. Spearman test was utilized for correlation analysis. P value <0.05 was considered statistically significant.
Results
Identification of Key AAA-Related m6A Regulators
Based on GSE47472 dataset, the distinct expression analysis of genes between AAA tissues and controls was conducted, as shown in the volcano plot of DEGs (Figure 1). And the expression pattern of m6A regulators is presented in Figure 2 and Table S1. The results revealed that three m6A regulators (METTL14, RBM15B and HNRNPC) were differentially expressed between AAA and control tissues. Specifically, the lower expression of METTL14 and HNRNPC and higher level of RBM15B were found in AAA tissues than those in normal aortas.
Potential Functions of the Modified Genes by AAA-Related m6A Regulators
The results of co-expression analysis showed that METTL14 expression was remarkably correlated with 4181 genes, and HNRNPC and RBM15B levels had a significant co-expression relationship with 13,355 and 8659 genes in AAA tissues, respectively. Table S2 shows the genes possibly modified by METTL14, HNRNPC and RBM15B through m6A2Target database. Thus, a total of 219, 383 and 1182 genes were separately regulated by METTL14, HNRNPC and RBM15B through m6A RNA modification in AAA disease (Table S3). Furtherly, GO and KEGG analyses were performed on these genes to interpret the clustering results from the perspective of fundamental biological processes. Finally, we observed a total of 429 GO terms and 7 KEGG pathways. The GO analysis showed that genes were mainly involved in RNA catabolic process, regulation of translation, protein targeting, ribosomal subunit, cell-substrate junction, focal adhesion, ubiquitin-like protein ligase binding, transcription coregulator activity, et al (Figure 3A and Table 2). In addition, three m6A regulators also participated in the regulation of immune response, glycolytic process, regulation of carbohydrate catabolic process, ATPase activity and regulation of autophagy (Figure 3A and Table 2). The KEGG results showed that these modified genes were enriched in ribosome, coronavirus diseases-COVID-19, RNA transport, protein processing in endoplasmic reticulum, cell cycle, et al (Figure 3B and Table 2).
 |
Table 2 The Major GO Terms and Significant KEGG Pathway Enrichment Analysis on the Modified Genes by METTL14, HNRNPC and RBM15B in AAA |
Association Between AAA-Related m6A Regulators and Immune Infiltration, Metabolism and Autophagy
Correlation analysis showed that the infiltration degree of Tcm, macrophages and mast cells was positively correlated with HNRNPC and METTL14 expression but negatively associated with RBM15B expression, whereas infiltrating Tgd and NK CD56bright cells had a negative relationship with HNRNPC and METTL14 but a positive link with RBM15B (Figure 4).
Moreover, in the target genes of METTL14, HNRNPC and RBM15B, a total of 154 genes were involved in metabolism and 76 genes were correlated with autophagy in AAA disease (Figure 5A and Table S4). Further PPI network analysis showed the interactive relationships among these metabolism or autophagy related genes (Figure 5B), and the top 20 hub genes were selected according to node degree (Table 3).
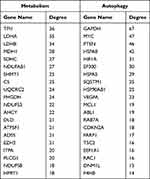 |
Table 3 List of the Top 20 Hub Genes Associated with Metabolism or Autophagy |
Construction of ceRNA Regulatory Network for AAA-Related m6A Regulators
The results of co-expression analysis showed that a total of 26, 96 and 62 lncRNAs were associated with METTL14, HNRNPC and RBM15B, respectively. After predicting miRNA-mRNA and lncRNA-miRNA interaction pairs, 16 miRNAs for METTL14, 9 miRNAs for HNRNPC and 8 miRNAs for RBM15B were identified in both interactions and then used for lncRNA-miRNA-mRNA network construction. The ceRNA regulatory networks for AAA-related m6A regulators are presented in Figure 6.
 |
Figure 6 Competing endogenous RNA (ceRNA) network for METTL14, HNRNPC and RBM15B. Notes: Orange color indicates m6A regulators, blue color indicates miRNAs, and green color indicates lncRNAs. |
Validation and Clinical Relevance of Key m6A Regulators in AAA
The baseline characteristics of subjects included in our study are presented in Table 4. Using qRT-PCR, we examined the mRNA expression of METTL14, HNRNPC and RBM15B in aortic tissues. METTL14 mRNA was confirmed to be significantly downregulated in AAA samples in comparison with control aortas (0.61±0.32 vs 1.11±0.62, P=0.040) (Figure 7A). When AAA cases were further classified into non-rupture and rupture groups, it could be observed that METTL14 was in lower expression in rupture cases compared to non-rupture group (0.34±0.19 vs 0.72±0.30, P=0.037) (Figure 7A). However, neither HNRNPC nor RBM15B expression exhibited a statistical difference between AAA and control group, although there was a trend toward down-regulation for HNRNPC (0.64±0.39 vs 1.09±0.54, P=0.076) and up-regulation for RBM15B (1.70±1.29 vs 1.21±0.72, P=0.353) in AAA tissues (Figure 7B and C).
 |
Table 4 Baseline Characteristics of Subjects Included in Our Study |
According to the median value of METTL14 mRNA level, AAA patients were further divided into low and high METTL14 expression groups. As shown in Table 5, AAA cases with low METTL14 expression had higher levels of WBC and CRP than those with high METTL14 expression, and the low expression of METTL14 mRNA was more likely to occur in ruptured AAA patients (all P<0.05).
 |
Table 5 Association Between Tissue mRNA Expression of METTL14 and Clinical Features in AAA Patients |
Furtherly, we verified METTL14 protein expression in serum specimens. Similar to the results of tissue mRNA expression, serum METTL14 levels were significantly lower in AAA cases and rupture group compared to controls (60.35±18.08 vs 72.63±19.58, P=0.014) and non-rupture group (53.23±12.95 vs 67.47±20.01, P=0.028), respectively (Figure 8A). In addition, we evaluated the association between serum METTL14 expression and clinical features in AAA patients, and found that serum METTL14 was negatively related to circulating WBC (r=−0.458, P=0.011) and CRP (r=−0.398, P=0.029) (Figure 8B and C).
Discussion
As a new dimension of gene expression regulation, m6A RNA methylation has attracted extensive attention and intense interest among the research community. In recent years, researchers have discovered that m6A regulators are of great importance in the occurrence, development and prognosis of kinds of human diseases. Although the pivotal role of m6A modification in cardiovascular disorders has been emphasized, study of m6A regulators in AAA field remains in its initial stage.19 To the best of our knowledge, this was the first systematic bioinformatics analysis of the landscape of m6A regulators in AAA disease, which could offer reliable directions for future-specific experimental researches of AAA and novel opportunities for developing effective therapies.
At first, the expression profile of common m6A regulators between AAA tissues and normal aortic tissues was described. The results based on public GEO data showed that two “writers” (METTL14 and RBM15B) and one “readers” (HNRNPC) had statistical significance, suggesting their possible functional importance in AAA. The m6A methyltransferases are responsible for catalyzing the formation of m6A modification.13 METTL14 serves as a supporting enzyme to strengthen the catalytic effect of m6A RNA methylation.13 RBM15B is the paralog of RBM15 and has been confirmed to bind METTL3 relying on the meditation of WTAP.20 As an RNA-binding protein of HNRNP family, HNRNPC is mainly located in cell nucleus and plays a regulatory role in the acceleration of pre-miRNA processing.21 Some researchers have verified the essential role of METTL14 in cardiac homeostasis, brain and immune system development, and skeletal function.6 Wang et al demonstrated that increased RBM15B expression was relevant to ovarian cancer cell metastases and HNRNPC was a predictor of paclitaxel resistance.22 However, Fang et al found that RBM15B expression was negatively correlated with multiple immune signatures in renal carcinoma,23 while Guo et al revealed that HNRNPC was abnormally up-regulated in lung cancer and related to worse outcomes.21
To further illustrate the downstream molecular mechanisms and functions involved in AAA-related m6A regulators, we selected a set of genes that might be regulated by METTL14, HNRNPC and RBM15B through m6A modification in AAA. The most enriched GO terms were RNA catabolic process, regulation of translation, ribosomal subunit, cell-substrate junction, focal adhesion, ubiquitin-like protein ligase binding, transcription coregulator activity, et al. The pathways enriched by KEGG were linked to ribosome, coronavirus diseases-COVID-19, RNA transport, cell cycle, et al. Some of these functions and pathways have been reported to accelerate the onset and development of AAA.24 For example, Zhang et al found that perturbed ribosomal DNA transcription and induction of nucleolar stress in SMCs that elicited cell cycle arrest could contribute to the pathogenesis of AAA,25 while Harada et al showed that focal adhesion kinase facilitated sustained aortic inflammation and AAA progression by modulating macrophage behavior.26 A recent study has reported that COVID-19 may theoretically influence AAA disease through multiple SARS-CoV-2-induced mechanisms, but whether and to what extent the COVID-19 pandemic will influence the prevalence, progression, and lethality of AAA is unclear.27
It was intriguing to note that above target genes were also enriched in biological processes linked with regulation of immune response, body metabolism and autophagy, which implied the connection between AAA-related m6A regulators and immunity, metabolism and autophagy. Studies have demonstrated that a host of immune cells can infiltrate into aortic walls of AAA, in which the roles of T lymphocytes and macrophages are predominantly studied.3 In AAA tissues, correlation analysis between AAA-related m6A regulator expression and infiltrating immune cell score suggested that METTL14 or HNRNPC downregulation and RBM15B upregulation might inhibit the infiltration of Tcm, macrophages and mast cells, and promote several immune cell aggregation, such as Tgd and NK CD56bright cells. This indicated the important and connecting role of abnormal METTL14, RBM15B and HNRNPC expression in epigenetic modification and immune infiltration implicated in AAA pathogenesis.
Currently, body metabolic disorders have been discovered to be closely associated with the occurrence and development of AAA by affecting the pathological processes of amino acid, lipid and carbohydrate metabolism in vivo.4 Qureshi et al demonstrated the utility of metabolic profiling in identifying potential AAA biomarkers and elucidating pathways underlying aneurysmal disease.28 In addition, there exists an associative link between dysregulated autophagy and human AAA, and growing evidence highlights the protective but dysfunctional role of autophagy in AAA formation.5,29 Loss of autophagy could promote VSMCs death and vascular inflammation, and aggravate AAA.30 Our analysis revealed a total of 154 metabolism-related genes and 76 autophagy-related genes modified by AAA-related m6A regulators, suggesting the possible biological functions of m6A regulators in AAA through modulating relevant gene expression in metabolism/autophagy pathway. Furtherly, PPI results showed that many of the modified genes involved in metabolism or autophagy had functional interactions, and hub genes were also selected. Some metabolic process-related genes, such as AHCY, CYP19A1, DNMT3A and EZH2, have been reported to participate in AAA.31–34 For identified autophagy-related genes, MCL1, HIF1A, VEGFA, PARP1, AFT4 and CANX are essential for the survival and function of vascular cells and underlie AAA.2,35–39 Meanwhile, recent studies proved that EZH2 expression was regulated through m6A modification in heat stress response and cancer progression,40,41 and VEGFA and PARP1 levels could be modulated in cancer cells in a m6A-dependent manner.42,43 However, whether m6A modification mediated by m6A regulators could influence the expression of essential metabolism/autophagy-related genes and thus contribute to AAA formation deserves further study.
The dysregulation of m6A regulator expression also has an important impact on the occurrence and progression of cardiovascular diseases.13 LncRNAs and miRNAs are a class of non‑coding RNAs, and lncRNAs can serve as competing endogenous RNA (ceRNA) and sponge miRNA to regulate the expression of a series of mRNAs.44 Based on this ceRNA theory, lncRNA-miRNA-mRNA networks for METTL14, HNRNPC and RBM15B were constructed through bioinformatics analysis to determine the interaction among the lncRNAs, miRNAs and mRNAs in AAA disease. Our results implied that ceRNA networks could provide novel clues for exploring the underlying regulatory mechanisms of key m6A regulators in the formation of AAA.
Furtherly, we detected the expression levels of METTL14, HNRNPC and RBM15B in our collected human samples, and found lower METTL14 expression in AAA subjects than that in controls. Interestingly, METTL14 was also markedly downregulated in rupture AAA group compared with non-rupture group. AAA cases with low METTL14 mRNA expression were prone to have higher levels of WBC and CRP. Similarly, serum protein expression of METTL14 displayed a negative association with circulating WBC and CRP in AAA patients. Our findings indicated that decreased METTL14 expression might be implicated in the disease progression and more severe inflammatory status, and METTL14 could be a promising biomarker and effective therapeutic target for AAA. However, He et al revealed no significant difference in the mRNA expression of METTL14 between human AAA tissues and healthy aortas,14 whereas Zhong et al found that METTL14 was significantly elevated in mouse AAA models.15 Some data revealed that METTL14 expression was downregulated in colorectal cancer, liver cancer, gastric cancer and bladder cancer, and reduced METTL14 was associated with poor overall survival.45–48 Tissue and cellular specificity for methyltransferase activation under certain conditions may account for these discrepancies.49
Several limitations should be acknowledged. First, our bioinformatics results were mainly based on the public chip data. Second, the sample size of our collected tissues was small, especially for normal aortas, which might be the reason why we found no significant differences in HNRNPC and RBM15B mRNA expression. Third, there was a lack of in vitro validation by molecular experiments. Further loss-of-function studies are necessary to elucidate the specific functions and mechanisms of m6A regulators in AAA disease.
Conclusions
We used a series of bioinformatics methods to investigate the gene expression features and potential functions of m6A regulators in human AAA, and try to construct downstream/upstream regulatory networks for key AAA-related m6A regulators. Further results verified by our clinical tissue and blood samples suggested that the low expression of METTL14 was connected with a high risk for AAA presence and its rupture. The present study paves the way for future researches with the aim to explore the pathogenic mechanisms and novel therapeutic strategies for AAA, targeting m6A regulators.
Data Sharing Statement
The data presented in this study are openly available in the NCBI Gene Expression Omnibus (GEO) repository (GSE47472).
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (82001828) and Key Laboratory of Cancer Intervention in Liaoning Province (LS201605).
Disclosure
The authors report no conflicts of interest in this work.
References
1. van de Luijtgaarden KM, Heijsman D, Maugeri A, et al. First genetic analysis of aneurysm genes in familial and sporadic abdominal aortic aneurysm. Hum Genet. 2015;134(8):881–893. doi:10.1007/s00439-015-1567-0
2. Liu Y, Wang X, Wang H, Hu T. Identification of key genes and pathways in abdominal aortic aneurysm by integrated bioinformatics analysis. J Int Med Res. 2020;48(4):300060519894437.
3. Yuan Z, Lu Y, Wei J, Wu J, Yang J, Cai Z. Abdominal aortic aneurysm: roles of inflammatory cells. Front Immunol. 2020;11:609161. doi:10.3389/fimmu.2020.609161
4. Hou Y, Guo W, Fan T, et al. Advanced research of abdominal aortic aneurysms on metabolism. Front Cardiovasc Med. 2021;8:630269. doi:10.3389/fcvm.2021.630269
5. Wang L, Liu S, Pan B, et al. The role of autophagy in abdominal aortic aneurysm: protective but dysfunctional. Cell Cycle. 2020;19(21):2749–2759. doi:10.1080/15384101.2020.1823731
6. He PC, He C. m(6) A RNA methylation: from mechanisms to therapeutic potential. EMBO J. 2021;40(3):e105977. doi:10.15252/embj.2020105977
7. Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74(4):640–650. doi:10.1016/j.molcel.2019.04.025
8. Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28(6):616–624. doi:10.1038/s41422-018-0040-8
9. Han Z, Yang B, Wang Q, Hu Y, Wu Y, Tian Z. Comprehensive analysis of the transcriptome-wide m(6)A methylome in invasive malignant pleomorphic adenoma. Cancer Cell Int. 2021;21(1):142. doi:10.1186/s12935-021-01839-6
10. Han J, Wang JZ, Yang X, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18(1):110. doi:10.1186/s12943-019-1036-9
11. Su Y, Huang J, Hu J. m(6)A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gastric cancer. Front Oncol. 2019;9:1038. doi:10.3389/fonc.2019.01038
12. Wang H, Xu B, Shi J. N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene. 2020;722:144076. doi:10.1016/j.gene.2019.144076
13. Qin Y, Li L, Luo E, et al. Role of m6A RNA methylation in cardiovascular disease (review). Int J Mol Med. 2020;46(6):1958–1972. doi:10.3892/ijmm.2020.4746
14. He Y, Xing J, Wang S, Xin S, Han Y, Zhang J. Increased m6A methylation level is associated with the progression of human abdominal aortic aneurysm. Ann Transl Med. 2019;7(24):797. doi:10.21037/atm.2019.12.65
15. Zhong L, He X, Song H, et al. METTL3 induces AAA development and progression by modulating N6-methyladenosine-dependent primary miR34a processing. Mol Ther Nucleic Acids. 2020;21:394–411. doi:10.1016/j.omtn.2020.06.005
16. Fang X, Duan SF, Gong YZ, Wang F, Chen XL. Identification of key genes associated with changes in the host response to severe burn shock: a bioinformatics analysis with data from the gene expression omnibus (GEO) database. J Inflamm Res. 2020;13:1029–1041. doi:10.2147/JIR.S282722
17. Deng S, Zhang H, Zhu K, et al. M6A2Target: a comprehensive database for targets of m6A writers, erasers and readers. Brief Bioinform. 2020;22(3):bbaa055.
18. Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–795. doi:10.1016/j.immuni.2013.10.003
19. Wu S, Zhang S, Wu X, Zhou X. m(6)A RNA methylation in cardiovascular diseases. Mol Ther. 2020;28(10):2111–2119. doi:10.1016/j.ymthe.2020.08.010
20. Patil DP, Chen CK, Pickering BF, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–373. doi:10.1038/nature19342
21. Guo W, Huai Q, Zhang G, et al. Elevated heterogeneous nuclear ribonucleoprotein C expression correlates with poor prognosis in patients with surgically resected lung adenocarcinoma. Front Oncol. 2020;10:598437. doi:10.3389/fonc.2020.598437
22. Wang Q, Zhang Q, Li Q, Zhang J, Zhang J. Clinicopathological and immunological characterization of RNA m(6) A methylation regulators in ovarian cancer. Mol Genet Genomic Med. 2021;9(1):e1547. doi:10.1002/mgg3.1547
23. Fang J, Hu M, Sun Y, Zhou S, Li H. Expression profile analysis of m6A RNA methylation regulators indicates they are immune signature associated and can predict survival in kidney renal cell carcinoma. DNA Cell Biol. 2020;39(12):2194–2211. doi:10.1089/dna.2020.5767
24. Li Z, Kong W. Cellular signaling in abdominal aortic aneurysm. Cell Signal. 2020;70:109575. doi:10.1016/j.cellsig.2020.109575
25. Zhang W, Cheng W, Parlato R, et al. Nucleolar stress induces a senescence-like phenotype in smooth muscle cells and promotes development of vascular degeneration. Aging (Albany NY). 2020;12(21):22174–22198.
26. Harada T, Yoshimura K, Yamashita O, et al. Focal adhesion kinase promotes the progression of aortic aneurysm by modulating macrophage behavior. Arterioscler Thromb Vasc Biol. 2017;37(1):156–165. doi:10.1161/ATVBAHA.116.308542
27. Xu B, Li G, Guo J, et al. Angiotensin-converting enzyme 2, coronavirus disease 2019, and abdominal aortic aneurysms. J Vasc Surg. 2021;
28. Qureshi MI, Greco M, Vorkas PA, Holmes E, Davies AH. Application of metabolic profiling to abdominal aortic aneurysm research. J Proteome Res. 2017;16(7):2325–2332. doi:10.1021/acs.jproteome.6b00894
29. Wu QY, Cheng Z, Zhou YZ, et al. A novel STAT3 inhibitor attenuates angiotensin II-induced abdominal aortic aneurysm progression in mice through modulating vascular inflammation and autophagy. Cell Death Dis. 2020;11(2):131. doi:10.1038/s41419-020-2326-2
30. Ramadan A, Singh KK, Quan A, et al. Loss of vascular smooth muscle cell autophagy exacerbates angiotensin II-associated aortic remodeling. J Vasc Surg. 2018;68(3):859–871. doi:10.1016/j.jvs.2017.08.086
31. Giusti B, Saracini C, Bolli P, et al. Genetic analysis of 56 polymorphisms in 17 genes involved in methionine metabolism in patients with abdominal aortic aneurysm. J Med Genet. 2008;45(11):721–730. doi:10.1136/jmg.2008.057851
32. Golledge J, Biros E, Warrington N, et al. A population-based study of polymorphisms in genes related to sex hormones and abdominal aortic aneurysm. Eur J Hum Genet. 2011;19(3):363–366. doi:10.1038/ejhg.2010.182
33. Legaki E, Klonaris C, Athanasiadis D, et al. DAB2IP expression in abdominal aortic aneurysm: EZH2 and mir-363-3p as potential mediators. In Vivo (Brooklyn). 2019;33(3):737–742. doi:10.21873/invivo.11533
34. Xia Q, Zhang J, Han Y, et al. Epigenetic regulation of regulatory T cells in patients with abdominal aortic aneurysm. FEBS Open Bio. 2019;9(6):1137–1143. doi:10.1002/2211-5463.12643
35. Wang W, Xu B, Xuan H, et al. Hypoxia-inducible factor 1 in clinical and experimental aortic aneurysm disease. J Vasc Surg. 2018;68(5):1538–1550e1532. doi:10.1016/j.jvs.2017.09.030
36. Ni XQ, Lu WW, Zhang JS, et al. Inhibition of endoplasmic reticulum stress by intermedin1-53 attenuates angiotensin II-induced abdominal aortic aneurysm in ApoE KO mice. Endocrine. 2018;62(1):90–106. doi:10.1007/s12020-018-1657-6
37. Xu B, Iida Y, Glover KJ, et al. Inhibition of VEGF (vascular endothelial growth factor)-A or its receptor activity suppresses experimental aneurysm progression in the aortic elastase infusion model. Arterioscler Thromb Vasc Biol. 2019;39(8):1652–1666. doi:10.1161/ATVBAHA.119.312497
38. Liang ES, Bai WW, Wang H, et al. PARP-1 (poly[ADP-ribose] polymerase 1) inhibition protects from ang II (angiotensin II)-induced abdominal aortic aneurysm in mice. Hypertension. 2018;72(5):1189–1199. doi:10.1161/HYPERTENSIONAHA.118.11184
39. Slusarz A, Pulakat L. The two faces of miR-29. J Cardiovasc Med (Hagerstown). 2015;16(7):480–490. doi:10.2459/JCM.0000000000000246
40. Chen WW, Qi JW, Hang Y, et al. Simvastatin is beneficial to lung cancer progression by inducing METTL3-induced m6A modification on EZH2 mRNA. Eur Rev Med Pharmacol Sci. 2020;24(8):4263–4270.
41. Kisliouk T, Rosenberg T, Ben-Nun O, Ruzal M, Meiri N. Early-life m6A RNA demethylation by fat mass and obesity-associated protein (FTO) influences resilience or vulnerability to heat stress later in life. eNeuro. 2020;7(3):
42. Dixit D, Prager BC, Gimple RC, et al. The RNA m6A reader YTHDF2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discov. 2021;11(2):480–499. doi:10.1158/2159-8290.CD-20-0331
43. Yang L, Chen Y, Liu N, et al. Low expression of TRAF3IP2-AS1 promotes progression of NONO-TFE3 translocation renal cell carcinoma by stimulating N(6)-methyladenosine of PARP1 mRNA and downregulating PTEN. J Hematol Oncol. 2021;14(1):46. doi:10.1186/s13045-021-01059-5
44. Tian L, Hu X, He Y, Wu Z, Li D, Zhang H. Construction of lncRNA-miRNA-mRNA networks reveals functional lncRNAs in abdominal aortic aneurysm. Exp Ther Med. 2018;16(5):3978–3986.
45. Liu X, Xiao M, Zhang L, et al. The m6A methyltransferase METTL14 inhibits the proliferation, migration, and invasion of gastric cancer by regulating the PI3K/AKT/mTOR signaling pathway. J Clin Lab Anal. 2021;35(3):e23655. doi:10.1002/jcla.23655
46. Gu C, Wang Z, Zhou N, et al. Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N(6)-methyladenosine of notch1. Mol Cancer. 2019;18(1):168. doi:10.1186/s12943-019-1084-1
47. Ma JZ, Yang F, Zhou CC, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65(2):529–543. doi:10.1002/hep.28885
48. Chen X, Xu M, Xu X, et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19(1):106. doi:10.1186/s12943-020-01220-7
49. Yang X, Zhang S, He C, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19(1):46. doi:10.1186/s12943-020-1146-4
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







