Back to Journals » International Journal of General Medicine » Volume 14
Expression and Prognostic Value Identification of Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2) in Brain Low-Grade Glioma
Authors Shi L, Zhang Q, Shou X, Niu H
Received 8 June 2021
Accepted for publication 23 July 2021
Published 14 August 2021 Volume 2021:14 Pages 4517—4527
DOI https://doi.org/10.2147/IJGM.S323858
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Lu-feng Shi,1 Qian Zhang,1 Xiao-ying Shou,2 Huan-jiang Niu1
1Department of Neurosurgery, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, 310016, Zhejiang, People’s Republic of China; 2Department of Nursing, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, 310016, Zhejiang, People’s Republic of China
Correspondence: Huan-jiang Niu
Department of Neurosurgery, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, 368 Xiasha Road, Hangzhou, Zhejiang, 310016, People’s Republic of China
Tel +86 13606618811
Email [email protected]
Objective: This study aimed to reveal the potential function of methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) and emphasized its importance in brain low-grade glioma (LGG).
Methods: We firstly explored the differential expression of MTHFD2 mRNA in LGG and normal tissues, followed by correlation analysis of MTHFD2 mRNA expression with patient’s clinical characteristics. MTHFD2 protein expression in LGG and subcellular location were also evaluated. Then, survival analysis was performed to reveal the influence of MTHFD2 expression on the overall survival of patients, and Cox regression analysis was applied to predict the prognostic factor for overall survival of LGG. Finally, we performed functional analysis to reveal potential MTHFD2-associated pathways involved in LGG.
Results: We found that MTHFD2 was highly expressed in LGG patients (P< 0.05), and MTHFD2 expression was related to patient’s age and IDH mutation status (all P< 0.05). MTHFD2 protein was mainly localized to the mitochondria. Survival analysis showed that high expression of MTHFD2 desirably improved the prognosis of LGG patients (P< 0.001), especially for those patients with age ≥ 45 years (P< 0.05). But independent prognostic role of MTHFD2 in LGG was not observed. Pathway enrichment analysis indicated that MTHFD2 high expression significantly and positively participated in the pathway of one carbon pool by folate (all P< 0.05).
Conclusion: High expression of MTHFD2 was observed in LGG, which was favorable for the overall survival of LGG patients. Our results assumed that MTHFD2 high expression might play a pivotal role in LGG through positively regulating pathway of one carbon pool by folate.
Keywords: MTHFD2, low-grade glioma, high expression, prognosis
Introduction
Glioma is the most common malignant tumor in the central nervous system, accounting for 70% of brain cancers.1 According to the latest statistic reports in China,2 101,600 new cases and 61,000 related deaths have appeared. The World Health Organization classified gliomas into grade 1 to grade 4 based on histology and clinical criteria.3 Brain low-grade gliomas (LGG), belonging to grade 2 tumor, accounted for 15–25% of all gliomas and included astrocytomas, oligoastrocytomas and oligodendrogliomas.4 LGG was relatively benign as well as slow-growing, and overall survival (OS) of children and adolescents with LGG exceeded 90% at 5 to 10 years.5 Despite growing slowly initially, essentially all LGG lesions eventually progress to high-grade glioma (HGG) (Grade 3/4).6,7 Approximately 50% of patients with LGGs will experience malignant transformation within 5 years.8 Previous study has identified several diagnostic and prognostic signatures, such as PWP2, FGR and BMP2, which may be utilized as biomarkers for diagnosis and prognosis, and for development of a treatment strategy for LGG.9 It was of great significance that identification of potential therapeutic biomarker for LGG patients to improve their clinical outcome.
Methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) is a mitochondrial enzyme involved in folate metabolism,10 as well as a bifunctional enzyme with methylene dehydrogenase and cyclohydrolase activities.11 Growing evidence demonstrated the up-regulation of MTHFD2 in various cancers and its over-expression was associated with tumor cell proliferation.12 Inhibition of MTHFD2 expression affected the proliferation of lung,13 breast14 and acute myeloid leukemia15 cancer cells. These studies also highlighted MTHFD2 as being a promising therapeutic target for cancer treatment. MTHFD2 was identified as the drug target to block breast cancer cell migration and invasion.16 Additionally, MTHFD2 has been suggested to play an important role and potentially be valuable in the prognosis and treatment of glioma.17 Furthermore, immunohistochemical validation revealed that MTHFD2 was positively expressed in diffuse LGG.18 Currently, it has been regarded as one of the candidate marker genes for tumor invasion, metastasis and poor prognosis.10 Despite the growing interest about MTHFD2 as a potential target in cancers, the potential function of this enzyme in LGG has not been defined.
The present study was aimed at revealing the potential role of MTHFD2 in LGG. We explored the MTHFD2 expression in LGG patients, assessed its influence on patient prognosis and revealed its prognostic value on overall survival. Finally, MTHFD2-associated pathway was explored to understand the potential mechanism involved in LGG.
Methods
MTHFD2 mRNA Expression Analysis
We firstly explored the mRNA expression profile of MTHFD2 across all tumor and normal tissues through Gepia database (http://gepia.cancer-pku.cn/). The differential expression of MTHFD2 mRNA in normal and LGG samples was also analyzed. Correlation analysis between MTHFD2 mRNA expression and patient’s age, BRAF genetic status, tumor locations, primary or recurrent tumor, and primary tumor that further progressed was performed using GSE60898 dataset, which was obtained from GEO database (https://www.ncbi.nlm.nih.gov/geo/). In addition, the effects of age, gender, tumor subtypes, IDH mutation status, pharmaceutical therapy, and radiation therapy on MTHFD2 expression was also explored using TCGA Lower Grade Glioma (LGG) cohort, which was obtained from UCSC Xena database (https://xenabrowser.net/). Due to the significance of MTHFD2 in brain cancer, we also explored the MTHFD2 mRNA expression in human brain by HPA database (https://www.proteinatlas.org/).
MTHFD2 Protein Expression
Furthermore, the protein expression of MTHFD2 in normal and LGG tissues was also evaluated in HPA. The subcellular location of MTHFD2 protein in cancer cells is determined by indirect immunofluorescence microscopy in human U2-OS cell line and mouse cell line NIH 3T3. Representative multi-color images showing the protein of interest in green are displayed. The images also include markers for the nucleus (blue), microtubules (red) and ER (yellow). As stated in database, all proteins are stained in U-2 OS for localizing the whole human proteome on a subcellular level in one specific cell line. In addition to the human cell lines, many proteins have been stained in the mouse cell line NIH 3T3, given that the human and mouse genes are orthologous.
MTHFD2 Prognosis Analysis
We then evaluated the influence of MTHFD2 expression on the overall survival of LGG patients by Gepia and TISIDB databases. For deeper understanding the influence of MTHFD2 expression on the patient’s survival, we used the clinical data of LGG patients from TCGA Lower Grade Glioma (LGG) cohort for conducting survival analysis. The LGG patients were divided into high- and low-expression groups according to the median of MTHFD2 expression. Subsequently, the influence of MTHFD2 expression on overall survival was analyzed based on age, gender, tumor subtypes, IDH mutation status, pharmaceutical therapy, and radiation therapy using Kaplan-Meier analysis and Log rank test. Moreover, Cox regression analysis was performed to predict the independent prognostic role of MTHFD2 for overall survival in LGG.
Functional Enrichment Analysis
In order to explore the potential function of MTHFD2, we firstly detected its co-expressed genes in LGG through cBioportal database. The genes with absolute correlation>0.5, P<0.001 and q<0.001 were selected for further functional analysis. We used BiNGO plug-in of Cytoscape to explore the enriched biological processes of these identified genes, and ClueGO plug-in to predict the associated KEGG pathways. Further, we performed the GSEA analysis to explore the potential mechanism of MTHFD2 in LGG. The analysis criteria for GSEA were set as follows: number of permutations (1000), permutations type (gene_set), enrichment statistic (weighted), metric for ranking genes (person), genes list ordering mode (descending). The terms with FDR<25% and P<0.05 were considered as significant pathways.
Interactions Analysis on MTHFD2 with Tumor-Immune System
We used the TISIDB to explore the association of MTHFD2 expression with immune subtypes and molecular subtypes in LGG. The immune subtypes include C1 (wound healing), C2 (IFN-gamma dominant), C3 (inflammatory), C4 (lymphocyte depleted), C5 (immunologically quiet) and C6 (TGF-b dominant). Molecular subtypes contain Classic-like, Codel, G-CIMP-high, G-CIMP-low, Mesenchymal-like and PA-like.
Statistical Analysis
All the statistical analyses were performed using SPSS 19.0. Independent t-test or Mann–Whitney U-test were performed for comparisons between the two groups. ANOVA or Kruskal‑Wallis test was performed for multiple comparisons followed by Bonferroni’s post‑hoc test. Survival analysis was performed by Kaplan-Meier method and Log rank test. P value less than 0.05 was regarded as statistical significance.
Results
Expression Analysis in LGG
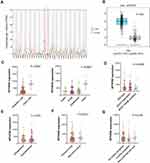
The MTHFD2 expression profile in human cancers (Figure 1A) presented that MTHFD2 was highly expressed in most cancer types including LGG, while down-regulation was observed in LAML (Acute Myeloid Leukemia) and THCA (Thyroid carcinoma) (all P<0.05). Differential expression analysis (Figure 1B) further confirmed high expression of MTHFD2 in LGG compared with normal group (P<0.05). Further, comparison analysis using GSE60898 dataset showed that age of LGG patient was associated with MTHFD2 expression, and higher MTHFD2 expression was observed in adult patient (Figure 1C, all P<0.001). However, no correlation was observed between MTHFD2 expression and BRAF genetic status, locations, primary or recurrent tumor, and primary tumor that further progressed (Figure 1D–G, all P>0.05).
Apart from above parameters, we also analyzed the correlation of MTHFD2 expression with other characteristics of patients using TCGA Lower Grade Glioma (LGG) cohort data. These samples just contained G2 and G3 patients, and this study has removed all G3 patients in the further investigations. The clinical characteristics of LGG patients in TCGA Lower Grade Glioma (LGG) cohort is presented in Table 1. Correlation in Figure 2A indicated that IDH mutation status of patient was related to MTHFD2 expression, and higher expression was observed in patients with IDH mutation (P=0.0260). Expression difference was not observed under different age, gender, tumor subtypes, pharmaceutical therapy, and radiation therapy (Figure 2B–F).
 |
Table 1 The Clinical Characteristics of LGG Patients in TCGA Lower Grade Glioma (LGG) Cohort |
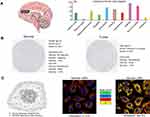
We then evaluated the MTHFD2 mRNA expression in the human brain. The results indicated that MTHFD2 was detected in all with a low region specificity (Figure 3A). Immunohistochemistry images (Figure 3B) showed that MTHFD2 protein was not detected in normal endothelial cells, but showed a weak intensity in normal glial cell. The MTHFD2 protein staining was not detected in cancer cell, but a weak intensity was observed. Subcellular location showed that MTHFD2 was mainly localized to the mitochondria both in human and mouse cancer cells (Figure 3C).
Prognosis Analysis in LGG
The effect of MTHFD2 expression on the clinical outcome of LGG patients was subsequently predicted (Figure 4). Survival analysis in Gepia suggested that MTHFD2 expression statistically correlated with the overall survival of patients, and MTHFD2 high expression prolonged the survival time of patients compared with low expression (P<0.001). The results from TISIDB also demonstrated the favorable impacts of MTHFD2 high expression on the overall survival of patients (P<0.001).
 |
Figure 4 The association of MTHFD2 expression with overall survival of LGG patients. |
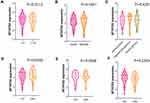
We then performed restrict survival analysis using TCGA Lower Grade Glioma (LGG) cohort data. The results indicated that expression of MTHFD2 was statistically related with overall survival time of patients with age ≥ 45 years (Table 2), and higher risk was observed in MTHFD2 low expression group (HR=4.530, P=0.040). However, survival difference between MTHFD2 high and low expression groups was not observed under other parameters (all P>0.05).
 |
Table 2 Restrict Survival Analysis on LGG Patients in TCGA Lower Grade Glioma (LGG) Cohort |
We also performed Cox regression analysis to predict the independent prognostic factor for overall survival of LGG patients. Although MTHFD2 was associated with survival of patients, its independent prognostic role in LGG was not detected in this study (Table 3).
 |
Table 3 Multivariate Cox Regression Analysis on LGG Patients |
Functional Enrichment Analysis
To further determine the potential function of MTHFD2 in LGG, we subsequently performed co-expression analysis on MTHFD2 through cBioportal database. A total of 20,161 co-expressed genes of MTHFD2 in LGG were found, and 175 genes with absolute value of correlation coefficient > 0.5 and P<0.001 were selected for further enrichment analysis.
We then explored the potential biological process associated with the 175 co-expressed genes by BiNGO of Cytoscape. The results (Figure 5) indicated that these co-expressed genes significantly participated in cellular process, cellular component organization, cell cycle, cell division, nuclear division, DNA replication and DNA metabolic process (all P<0.001).
 |
Figure 5 The enriched biological process of 175 co-expressed genes with MTHFD2. |
The potential KEGG pathways associated with 175 genes were explored through ClueGO of Cytoscape. Pathway analysis in Figure 6A showed that most of these genes participated in Oocyte meiosis (20.0%) and Fanconi anemia pathway (20.0%). Another important pathway detected included Cellular senescence (10.0%), p53 signaling pathway (10.0%), Cell cycle (10.0%), DNA replication (10.0%), One carbon pool by folate (10.0%) and Pyrimidine metabolism (10.0%). Among these pathways, MTHFD2 statistically referred to the pathway of One carbon pool by folate (Figure 6B).
 |
Figure 6 Potential enriched KEGG pathways of 175 co-expressed genes with MTHFD2. (A) The top 8 significant pathways. **P<0.01. (B) Interaction between pathway and related genes. |
We further explored the potential mechanism associated with MTHFD2 in LGG through GSEA analysis. The GSEA analysis totally detected 10 positive and 19 negative MTHFD2-associated pathways involved in LGG. According to GSEA analysis, we found that MTHFD2 high expression was statistically and positively related to One carbon pool by folate pathway (Figure 7A, P=0.0181). The detailed information about One carbon pool by folate pathway is presented in Figure 7B.
Tumor-Immune System Interaction Analysis on MTHFD2
We finally explored the interaction of MTHFD2 with tumor-immune system in TISIDB. The results indicated that MTHFD2 was statistically associated with immune subtypes in LGG (Figure 8A, P<0.001), and higher expression was observed in C5 (immunologically quiet) subtype. In addition, molecular subtypes in LGG also affected the MTHFD2 expression (Figure 8B, P<0.001), and Codel subtype showed a higher expression.
Discussion
Previous study has showed that MTHFD2 expression is low or absent in a large panel of normal tissues, and expressed in the developing embryo but absent in most healthy adult tissues, even those that are proliferating.19 MTHFD2 has been proved to be highly expressed in human cancers and associated with the tumor progression. Previous research indicated that MTHFD2 may serve as a tumor promoter by promoting cell proliferation. For example, MTHFD2 was found highly expressed in colorectal cancer (CRC) cells, and silence of MTHFD2 expression inhibited the proliferation, weakened the migration ability and promoted the apoptosis of CRC cells.20 MTHFD2 was also significantly over-expressed in non-small cell lung cancer (NSCLC) tissues, and knockdown of MTHFD2 resulted in reduction of tumor cell growth and tumorigenicity.21 In addition, MTHFD2 over-expression was associated with cell proliferation and vimentin-modulated migration and invasion in renal cell carcinoma (RCC).22 Moreover, MTHFD2 expression was significantly associated with the clinical outcome of patients, and high expression of MTHFD2 resulted in poor prognosis in hepatocellular carcinoma,23 esophageal squamous cell carcinoma24 and breast cancer.25 Taken together, MTHFD2 was oncogenic in nature and may serve as a prognostic indicator.
In this study, we found that MTHFD2 mRNA was over-regulated in LGG patients compared with normal group. Among LGG patients, higher expression was observed in adult patients. However, survival analysis indicated that MTHFD2 high expression was conducive to prognosis of LGG patients, especially facilitated the clinical outcome of patients with age ≥45 years. It seemed that MTHFD2 high expression exerted a positive role in LGG. The impact of MTHFD2 on LGG was different from that in other cancer types. We speculated that function of MTHFD2 exerted cancer type specificity.
It should be noted that previous findings about MTHFD2 in brain glioma seemed to be controversial. A bioinformatics analysis17 by Liu et al found that MTHFD2 was identified as a down-regulated gene in glioma, and the down-regulation of MTHFD2 was closely associated with a decreased overall survival among patients with glioma. However, Xu et al analyzed the expression of MTHFD2 in glioma tissues through TCGA analysis and qRT-PCR, finding that expression of MTHFD2 was significantly greater in glioma tissues than in normal tissues, and MTHFD2 was a protected factor for glioma patients as high expression with a long survival time.26 Gao et al demonstrated that MTHFD2 significantly correlated with overall survival, and it was a protected factor of glioblastoma multiforme patients.27 Our analysis also indicated the up-regulation of MTHFD2 in LGG, and MTHFD2 high expression was favorable to the overall survival of LGG patients. According to our investigation, the subjects in Liu et al and Xu et al studies were all patients with high-grade glioma, but the result of MTHFD2 expression analysis were opposite. Current study contained patient with low-grade glioma in G2, expression analysis indicated its up-regulation which agreed with Xu et al study. Although differences were observed about MTHFD2 expression among existing research in brain glioma, the assessment on prognostic value of MTHFD2 seemed to be consistent. All study suggested that MTHFD2 high expression was conducive to the prognosis of patients with glioma compared with low expression. The causes and mechanisms behind the discrepant expression profiles remained elusive and deserved further investigations.
We also performed pathway enrichment analysis to explore the potential MTHFD2-associated mechanism involved in LGG. Both KEGG and GSEA analysis indicated that MTHFD2 mainly participated in pathway of one carbon pool by folate. Metabolic pathway was the common mechanism related to MTHFD2. Folate-mediated one-carbon metabolism plays a central role in a broad array of metabolic processes required for the survival and growth of tumor cells.28 The patients with aminopterin treatment, an intermediate of folate metabolism, showed clinical, hematologic and pathological evidence of improvement.29 Metabolism reprogramming was one of the most active fields in cancer research, and the effects and consequences of elevated MTHFD2 in brain low-grade glioma were largely unknown. Thus, experiment and clinical studies were needed to understand its potential functional role in brain glioma.
In addition, recent studies have highlighted the critical value of MTHFD2 as a therapeutic target for treatment of cancers. Xu et al suggested that microRNA-940 could inhibit glioma progression by blocking mitochondrial folate metabolism through targeting of MTHFD2.26 Yam et al indicated that microRNA-33a-5p suppressed the growth and migration of colorectal cancer cell by inhibiting MTHFD2.30 Li et al reported that upregulation of miR-504-3p was associated with favorable prognosis of acute myeloid leukemia and may serve as a tumor suppressor by targeting MTHFD2.31 Moreover, folate analog as MTHFD2 inhibitor displayed therapeutic activity against colorectal cancer.32 These studies implied the significance of MTHFD2 as an effective target for more cancer treatment in the future.
Conclusions
This current study demonstrated that mRNA expression of MTHFD2 was significantly increased in LGG, and associated with age, and TP53-mutation status of patients. High expression of MTHFD2 favorably prolonged the patient’s survival time, and MTHFD2 was determined as an independent prognostic biomarker for overall survival of LGG patients. We further demonstrated that MTHFD2 was positively implicated in the pathway of one carbon pool by folate. Our research initially emphasized the importance of MTHFD2 in LGG, and the experimental intervention must be icing on the cake for this research.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Funding
This work was supported by the Zhejiang Provincial Department of Health Project (No. 201466094).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2014;16(Suppl 4):iv1–iv63. doi:10.1093/neuonc/nou223
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi:10.3322/caac.21338
3. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi:10.1007/s00401-016-1545-1
4. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi:10.1007/s00401-007-0243-4
5. Gnekow AK, Walker DA, Kandels D, et al. A European randomised controlled trial of the addition of etoposide to standard vincristine and carboplatin induction as part of an 18-month treatment programme for childhood (</=16 years) low grade glioma - A final report. Eur J Cancer. 2017;81:206–225.
6. Forst DA, Nahed BV, Loeffler JS, Batchelor TT. Low-grade gliomas. Oncologist. 2014;19:403–413. doi:10.1634/theoncologist.2013-0345
7. Morshed RA, Young JS, Hervey-Jumper SL, Berger MS. The management of low-grade gliomas in adults. J Neurosurg Sci. 2019;63:450–457. doi:10.23736/S0390-5616.19.04701-5
8. Afra D, Osztie E, Sipos L, Vitanovics D. Preoperative history and postoperative survival of supratentorial low-grade astrocytomas. Br J Neurosurg. 1999;13:299–305. doi:10.1080/02688699943727
9. Islam T, Rahman MR, Shuvo MA, Shahjaman M, Islam MR, Karim MR. Drug repositioning and biomarkers in low-grade glioma via bioinformatics approach. Inform Med Unlocked. 2019;17:100250. doi:10.1016/j.imu.2019.100250
10. Tibbetts AS, Appling DR. Compartmentalization of mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr. 2010;30:57–81.
11. Gustafsson R, Jemth AS, Gustafsson NM, et al. Crystal structure of the emerging cancer target MTHFD2 in complex with a substrate-based inhibitor. Cancer Res. 2017;77:937–948. doi:10.1158/0008-5472.CAN-16-1476
12. Tedeschi PM, Vazquez A, Kerrigan JE, Bertino JR. Mitochondrial Methylenetetrahydrofolate Dehydrogenase (MTHFD2) overexpression is associated with tumor cell proliferation and is a novel target for drug development. Mol Cancer Res. 2015;13:1361–1366. doi:10.1158/1541-7786.MCR-15-0117
13. Nishimura T, Nakata A, Chen X, et al. Cancer stem-like properties and gefitinib resistance are dependent on purine synthetic metabolism mediated by the mitochondrial enzyme MTHFD2. Oncogene. 2019;38:2464–2481. doi:10.1038/s41388-018-0589-1
14. Koufaris C, Valbuena GN, Pomyen Y, et al. Systematic integration of molecular profiles identifies miR-22 as a regulator of lipid and folate metabolism in breast cancer cells. Oncogene. 2016;35(21):2766–2776. doi:10.1038/onc.2015.333
15. Pikman Y, Puissant A, Alexe G, et al. Targeting MTHFD2 in acute myeloid leukemia. J Exp Med. 2016;213:1285–1306. doi:10.1084/jem.20151574
16. Lehtinen L, Ketola K, Makela R, et al. High-throughput RNAi screening for novel modulators of vimentin expression identifies MTHFD2 as a regulator of breast cancer cell migration and invasion. Oncotarget. 2013;4:48–63. doi:10.18632/oncotarget.756
17. Liu M, Xu Z, Du Z, et al. The identification of key genes and pathways in glioma by bioinformatics analysis. J Immunol Res. 2017;2017:1–9.
18. Wang S, Jin F, Fan W. Gene expression meta-analysis in diffuse low-grade glioma and the corresponding histological subtypes. Sci Rep. 2017;7:11741. doi:10.1038/s41598-017-12087-y
19. Nilsson R, Jain M, Madhusudhan N. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun. 2014;5:3128. doi:10.1038/ncomms4128
20. Wei Y, Liu P, Li Q, et al. The effect of MTHFD2 on the proliferation and migration of colorectal cancer cell lines. Onco Targets Ther. 2019;12:6361–6370. doi:10.2147/OTT.S210800
21. Yu C, Yang L, Cai M, et al. Down-regulation of MTHFD2 inhibits NSCLC progression by suppressing cycle-related genes. J Cell Mol Med. 2020;24(2):1568–1577. doi:10.1111/jcmm.14844
22. Lin H, Huang B, Wang H, et al. MTHFD2 overexpression predicts poor prognosis in renal cell carcinoma and is associated with cell proliferation and vimentin-modulated migration and invasion. Cell Physiol Biochem. 2018;51(2):991–1000. doi:10.1159/000495402
23. Liu X, Huang Y, Jiang C, et al. Methylenetetrahydrofolate dehydrogenase 2 overexpression is associated with tumor aggressiveness and poor prognosis in hepatocellular carcinoma. Dig Liver Dis. 2016;48(8):953–960. doi:10.1016/j.dld.2016.04.015
24. He H, Li P-C, Jia W, Hu B, Ji C-S. High expression of Methylenetetrahydrofolate Dehydrogenase 2 (MTHFD2) in Esophageal squamous cell carcinoma and its clinical prognostic significance. Med Sci Monit. 2020;26:e920259. doi:10.12659/MSM.920259
25. Liu F, Liu Y, He C, et al. Increased MTHFD2 expression is associated with poor prognosis in breast cancer. Tumour Biol. 2014;35(9):8685–8690. doi:10.1007/s13277-014-2111-x
26. Xu T, Zhang K, Shi J, et al. MicroRNA-940 inhibits glioma progression by blocking mitochondrial folate metabolism through targeting of MTHFD2. Am J Cancer Res. 2019;9:250–269.
27. Gao WZ, Guo LM, Xu TQ, Yin YH, Jia F. Identification of a multidimensional transcriptome signature for survival prediction of postoperative glioblastoma multiforme patients. J Transl Med. 2018;16:368. doi:10.1186/s12967-018-1744-8
28. Green NH, Galvan DL, Badal SS, et al. MTHFD2 links RNA methylation to metabolic reprogramming in renal cell carcinoma. Oncogene. 2019;38(34):6211–6225. doi:10.1038/s41388-019-0869-4
29. Farber S, Diamond LK, Mercer RD, Sylvester RF, Wolff JA. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid (aminopterin). N Engl J Med. 1948;238(23):787–793. doi:10.1056/NEJM194806032382301
30. Yan Y, Zhang D, Lei T, et al. MicroRNA-33a-5p suppresses colorectal cancer cell growth by inhibiting MTHFD2. Clin Exp Pharmacol Physiol. 2019;46:928–936. doi:10.1111/1440-1681.13125
31. Li SM, Zhao YQ, Hao YL, Liang YY. Upregulation of miR-504-3p is associated with favorable prognosis of acute myeloid leukemia and may serve as a tumor suppressor by targeting MTHFD2. Eur Rev Med Pharmacol Sci. 2019;23:1203–1213.
32. Ju HQ, Lu YX, Chen DL. Modulation of redox homeostasis by inhibition of MTHFD2 in colorectal cancer: mechanisms and therapeutic implications. J Natl Cancer Inst. 2019;111:584–596. doi:10.1093/jnci/djy160
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.