Back to Journals » Cancer Management and Research » Volume 12
Expression and Prognostic Significance of BANF1 in Triple-Negative Breast Cancer
Authors Zhang G
Received 28 August 2019
Accepted for publication 17 December 2019
Published 8 January 2020 Volume 2020:12 Pages 145—150
DOI https://doi.org/10.2147/CMAR.S229022
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Genhao Zhang
The Department of Blood Transfusion, Zhengzhou University First Affiliated Hospital, Zhengzhou, People’s Republic of China
Correspondence: Genhao Zhang
Blood Transfusion, Zhengzhou University First Affiliated Hospital, Zhengzhou, People’s Republic of China
Tel +86 158 3816 5680
Email [email protected]
Aim: To investigate the expression of barrier-to-autointegration factor 1 (BANF1) and its prognostic significance in triple-negative breast cancer (TNBC).
Methods: BANF1 immunohistochemical detection was performed in 60 TNBC specimens and 30 normal control tissues. Real-time PCR was performed to assess the expression of BANF1 gene in TNBC tissues and their correlations with proliferation and metastasis. Kaplan-Meier survival analysis was used to assess the effect of BANF1 expression on the relapse-free survival (RFS) of TNBC patients. Univariable and multivariable Cox proportional hazards regression model analysis was used to confirm independent prognostic factors.
Results: Expression of BANF1 in TNBC was significantly higher than that of the normal control group (p<0.001), and it was related to the status of lymph node metastasis and TNM staging (p<0.05), and not related to age and tumor size (p>0.05). BANF1 expression has a positive correlation with MKI67 and MTA1 expression (p<0.01). Univariable analysis showed that expression of BANF1, the status of lymph node metastasis and TNM stage were related to the relapse-free survival (RSF) of TNBC patients (p<0.001, p=0.001, p=0.013, respectively). Multivariable Cox regression indicated that the status of lymph node metastasis was an independent prognostic factor for TNBC patients (p<0.001). The survival curve suggested that the survival times for TNBC patients with high BANF1 expression have no difference compared with that for the low-expression patients (p>0.05).
Conclusion: Expression of BANF1 may play a role in the occurrence and development of TNBC. Lymph node metastasis was the only independent prognostic factor predicts a poor prognosis.
Keywords: BANF1, relapse-free survival, prognosis
Introduction
Triple-negative breast cancer (TNBC) is a highly aggressive form of breast cancer that lacks targeted therapy options, which lacks estrogen receptor (ER) and progesterone receptor (PR) expression and are negative for human epidermal growth factor receptor 2 (HER2) overexpression;1 moreover, TNBC does not respond to hormonal or anti-HER2 therapies and currently lacks targeted therapy options. Patients diagnosed with TNBC after chemotherapy have poorer outcomes than patients with other breast cancer subtypes.2 Barrier-to-autointegration factor 1 (BANF1) is a highly conserved DNA-binding protein that forms homodimers and has a variety of functions associated with the maintenance of the intact cellular genome, which regulates gene expression, participates in the formation of karyotin structures and is associated with cell mitosis,3 indicating its vital role in the process of malignant transformation of cells. The present study was designed to investigate the expression profile of BANF1 in TNBC and its relationship with clinical-pathological characteristics and to explore the relationship between BANF1 and the prognosis of patients with TNBC by survival analysis.
Materials and Methods
Clinical Data
Sixty TNBC specimens and 30 corresponding non-cancerous tissues (normal tissues) from patients admitted to the Department of Pathology of the First Hospital of Zhengzhou University from 2012 to 2013 were selected. None of the patients were treated with radiotherapy or chemotherapy before surgery and those with incomplete data were excluded. Patients enrolled were approved by the ethics committee of the First Affiliated Hospital of Zhengzhou University. All pathological data were reviewed and joint diagnoses were made by two senior pathologists. Follow-up data were available for all patients up to January 2017, with a follow-up time ranging from 1 to 60 months. Of the patients, 35 survived, 21 died and 4 were unknown.
Methods
Immunohistochemistry was performed to assess BANF1 expression in TNBC and non-cancerous tissues. Paraffin-embedded breast tissue samples were cut at a thickness of 5 mm and then mounted on coated microscope slides. Briefly, antigen retrieval was conducted via immersion of the slides in the citrate-EDTA buffer, followed by heating in a microwave oven for 2 min at high power and 20 min at low power. Non-specific staining was blocked using 5% goat serum. After blocking, 50 mL of the primary antibody (BANF1) was applied to each section overnight at 4°C. A mouse IgG isotype control antibody was used at the same concentration as the primary antibodies. On the day after incubation with the secondary antibody, sections were incubated with DAB until the desired staining developed. Interpretation of immunohistochemical results Microscopic results revealed that BANF1 protein was expressed in the nucleus of tumor cells. A count of positive-stained cells was performed and staining intensity was observed, and the percentage of positive cells was calculated (negative=0, 1–10% of positive cells=1, 11–50%=2, 51–80%=3, 81–100%=4) and the staining intensity of positive cells was determined (negative=0, weak positive=1, positive=2, strong positive=3). The product of the percentage and the intensity was used to determine the level of expression: ≤4 was an indicator of low expression or no expression and >4 as high expression.
Real-time PCR was performed to assess the expression of BANF1 gene in TNBC and their correlations with proliferation and metastasis. The total RNA was extracted using TRIzol reagent from tissues and then reverse-transcribed using a PrimeScript RT Master Mix Synthesis Kit for mRNAs. The relative expression levels of mRNAs were quantified using Maxima SYBR Green qRT-PCR Master Mix (2×) in a Real-Time PCR system. The expression level of β-ACTIN was used as endogenous controls, and the fold changes were calculated using the 2−ΔΔct method. The primer sequences are listed as follows: BANF1, F: TGGCTGAAAGACACTTGTGG, R: CACTCTCGAAGGCATCCGAAG; MKI67, F: ACAACAGGAGGAGGAAGTGCT, R: TTGACCTAGACGCGGGGAT; MTA1, F: AGCTACGAGCAGCACAACGGGGT, R: CACGCTTGGTTTCCGAGGAT and β-ACTIN, F: CGTGGGCCGCCCTAGGCA CCA, R: TTGGCTTAGGGTTCAGGGGGG.
Statistical Analysis
Statistical analyses were performed using Statistical Package for the Social Sciences (SPSS 22.0). Categorical data were compared with Pearson chi-square test or Fisher exact test whenever appropriate, and continuous variables were compared with independent-samples t-test or Mann–Whitney U-test. Kaplan-Meier survival analysis, with the log-rank test, was used to assess the effect of BANF1 expression on the relapse-free survival (RFS) of TNBC patients. Univariable and multivariable Cox proportional hazards regression model was used to analyze the association between the clinical factors and overall survival. A P value <0.05 was considered statistically significant.
Results
Difference in Expression of BANF1 Protein Between TNBC and Normal Tissues
BANF1 was expressed in 49 cases of the 60 TNBC tissues to a certain degree (Figure 1), and 5 in cases of the 30 normal tissues with weakly positive expression. The corresponding rates were 81.6% and 16.6%. The difference between the two groups was statistically significant (χ2 = 35.208, p<0.001).
 |
Figure 1 Expression of BANF1 protein in TNBC tissues. (A) Low expression. (B) High expression. EnVision ×200. |
Relationship Between Over-Expressed BANF1 and Clinical Pathological Data in TNBC
Table 1 shows that the expression of BANF1 protein was related to lymph node metastasis and TNM staging (p<0.05), not related to age and tumor size (p>0.05).
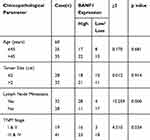 |
Table 1 Relationship Between BANF1 Expression and Clinical Pathological Features |
Relationship Between BANF1 and Marker of Proliferation and Metastasis
Figure 2 shows that BANF1 mRNA was overexpressed in TNBC, and had a positive correlation with markers of proliferation and metastasis (MKI67 and MTA1, all p <0.05).
Survival Analysis
The survival curve suggested that the survival times for TNBC patients with high BANF1 expression have no difference compared with that for the low-expression patients (p>0.05, Figure 3A). To supplement the observation made with our patient dataset, evaluation of the relationship of gene expression and survival was carried out with publicly available datasets using KM Plotter.4 The results indicated no significant differences between high and low BANF1 gene expression on relapse-free survival (RFS) of TNBC patients (p>0.05, Figure 3B). Subsequently, the survival of patients with TNBC correlates with the expression of BANF1, lymph node metastasis and TNM stage (p<0.05) by univariable analysis; however, lymph node metastasis was the only independent prognostic factors for patients with TNBC by multivariable Cox proportional regression model (p<0.05, Table 2).
 |
Table 2 Univariable and Multivariate Cox Regression Analysis for Screening Prognostic Factors |
Discussion
TNBC has an aggressive nature, and patients diagnosed with TNBC lack targeted therapy options.5 Surgical removal of the primary tumor is a common practice in TNBC treatment. However, postsurgical metastasis poses an immense setback in cancer therapy. Thus, the development of potential therapeutic targets is urgently needed. In recent years, an increasing number of studies had focused on understanding the molecular and biological characteristics of TNBC. In this study, we investigated the expression of BANF1 in TNBC and explored the effects of prognostic significance in TNBC tissues. We found that BANF1 was over-expressed in TNBC tissues and related to lymph node metastasis and TNM staging, indicating that expression of BANf1 may be one of the main causes of the occurrence and development of TNBC. However, our survival analysis results suggested that no significant differences between high and low BANF1 gene expression on relapse-free survival (RFS) of TNBC patients, which indicates that BANF1 has an impact on prognosis together with other factors, but is not an independent prognostic factor by itself.
Members of the BAF complex family have a variety of functions associated with the maintenance of the intact cellular genome, which are reported to play essential roles in carcinogenesis and cancer progression.6,7 As an important part of the lamina, BANF1 encodes a highly conserved BAF protein consisting of 89 amino acids and binding to double-stranded DNA with high-affinity. BANF1 is located at the core of chromatin and participate in completing the nuclear membrane reformation, indicating that BANF1 plays an important role in cell mitosis.8 We can regulate numerous cellular activities, including protein dimerization, its binding to DNA, and subcellular localization of the protein, via regulating of BAF phosphorylation.9 It has been reported that BANF1 may be a target for the treatment of tumors with a specialized inhibitor aimed at BAF phosphorylation mediated by vaccinia-related kinase 1.10 Shen has reported that BANF1 was an early-stage cancer decision markers and drivers of hepatocellular carcinoma.11 In addition, Li reported that BANF1 expression is increased in gastric cancer and associated with the poor prognosis of patients, which was a novel biomarker for gastric cancer.12
Our experimental results showed that the expression of BANF1 was significantly increased in TNBC tissues, indicating that BANF1 plays an important role in the carcinogenesis of TNBC. BANF1 protein was not only highly expressed in TNBC, but also related to the clinical-pathological parameter, such as the status of lymph node metastasis and TNM stage, which indicated that the progression of TNBC may be related to the expression of BANf1. What is more, BANF1 mRNA was overexpressed in TNBC and had a positive correlation with markers of proliferation and metastasis (MKI67 and MTA1). Next, the results of COX regression analysis suggested that the status of lymph node metastasis was an independent risk factor affecting the RSF of patients with TNBC, which indicates that BANF1 has an impact on prognosis together with other factors, but is not an independent prognostic factor by itself. A limitation of the present study was that the BANF1 protein levels in the tissues from patients with TNBC were only detected by immunohistochemistry and that Western blot analysis is required to support the immunohistochemistry results. Further, our study is a cohort with small sample sizes. Data from future well-designed studies involving multiple centers and large numbers of patients will be required to provide vigorous assessment and validation of the efficacy of our conclusions.
In summary, BANF1 was over-expressed in clinical TNBC tissues, and was associated with the status of lymph node metastasis and TNM stage. Additionally, the results of the present study confirmed that BANF1 probably correlates with the occurrence and development of TNBC and might be a potential molecular therapeutic target for TNBC.
Ethics Approval and Consent to Participate
The present study was approved by the Ethics Committees of the First Affiliated Hospital of Zhengzhou University. Written informed consent was obtained from all patients.
Disclosure
The author reports no conflicts of interest in this work.
References
1. Nakhjavani M, Palethorpe HM, Tomita Y, et al. Stereoselective anti-cancer activities of ginsenoside Rg3 on triple negative breast cancer cell models. Pharmaceuticals (Basel). 2019;12(3). doi:10.3390/ph12030117
2. Mazumdar A, Tahaney WM, Reddy Bollu L, et al. The phosphatase PPM1A inhibits triple negative breast cancer growth by blocking cell cycle progression. NPJ Breast Cancer. 2019;5:22. doi:10.1038/s41523-019-0118-6
3. Dharmaraj T, Guan Y, Liu J, Badens C, Gaborit B, Wilson KL. Rare BANF1 alleles and relatively frequent EMD alleles including ‘healthy lipid’ emerin p.D149H in the ExAC cohort. Front Cell Dev Biol. 2019;7:48. doi:10.3389/fcell.2019.00048
4. Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi:10.1007/s10549-009-0674-9
5. Srinivasalu VK, Susan A, Jose WM, Philip A, Nambiar A, Pavithran K. 41P prognostic significance of BCL-2 expression in triple negative breast cancer (TNBC). Ann Oncol. 2019;30(Supplement_3):mdz095–040
6. Wiebe MS, Jamin A, Pfeiffer J. The barrier to autointegration factor: interlocking antiviral defense with genome maintenance. J Virol. 2016;90(8):3806–3809. doi:10.1128/JVI.00178-16
7. Molitor TP, Traktman P, Hetzer M. Depletion of the protein kinase VRK1 disrupts nuclear envelope morphology and leads to BAF retention on mitotic chromosomes. Mol Biol Cell. 2014;25(6):891–903. doi:10.1091/mbc.e13-10-0603
8. Brachner A, Braun J, Ghodgaonkar M, et al. The endonuclease Ankle1 requires its LEM and GIY-YIG motifs for DNA cleavage in vivo. J Cell Sci. 2012;125(Pt 4):1048–1057. doi:10.1242/jcs.098392
9. Birendra K, May DG, Benson BV, et al. VRK2A is an A-type lamin-dependent nuclear envelope kinase that phosphorylates BAF. Mol Biol Cell. 2017;28(17):2241–2250. doi:10.1091/mbc.e17-03-0138
10. Li J, Wang T, Pei L, et al. Expression of VRK1 and the downstream gene BANF1 in esophageal cancer. Biomed Pharmacother. 2017;89:1086–1091. doi:10.1016/j.biopha.2017.02.095
11. Shen Q, Eun JW, Lee K, et al. Barrier to autointegration factor 1, procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3, and splicing factor 3b subunit 4 as early-stage cancer decision markers and drivers of hepatocellular carcinoma. Hepatology. 2018;67(4):1360–1377. doi:10.1002/hep.v67.4
12. Li J, Hu B, Fang L, et al. Barrier-to-autointegration factor 1: a novel biomarker for gastric cancer. Oncol Lett. 2018;16(5):6488–6494. doi:10.3892/ol.2018.9432
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


