Back to Journals » Journal of Inflammation Research » Volume 11
Expression analysis of protein inhibitor of activated STAT (PIAS) genes in IFNβ-treated multiple sclerosis patients
Authors Taheri M , Azimi G , Sayad A, Mazdeh M, Arsang-Jang S, Omrani MD , Ghafouri-Fard S
Received 13 September 2018
Accepted for publication 8 November 2018
Published 6 December 2018 Volume 2018:11 Pages 457—463
DOI https://doi.org/10.2147/JIR.S187414
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Mohammad Taheri,1,* Ghazaleh Azimi,2,* Arezou Sayad,2 Mehrdokht Mazdeh,3 Shahram Arsang-Jang,4,5 Mir Davood Omrani,5 Soudeh Ghafori-Fard2
1Student Research Committee, Shahid Beheshti University of Medical Sciences, Tehran, Iran; 2Department of Medical Genetics, Shahid Beheshti University of Medical Sciences, Tehran, Iran; 3Neurophysiology Research Center, Hamadan University Medical Sciences, Hamadan, Iran; 4Clinical Research Development Center (CRDU), Qom University of Medical Sciences, Qom, Iran; 5Urogenital Stem Cell Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
*These authors contributed equally to this work
Objectives: Protein inhibitors of activated STAT (PIAS) are transcription co-regulator of the Janus kinase/signal transducer and activator of transcription signaling pathway as well as nuclear factor-κB family of transcription factors. Both of them are involved in cytokine release during inflammatory response.
Patients and methods: Considering the role of cytokine imbalance in the pathogenesis of multiple sclerosis (MS), we compared blood expressions of PIAS1-4 genes in 48 interferon β (IFNβ)-treated MS patients with those of healthy subjects by means of real time PCR.
Results: Although the expression levels of these genes were not significantly different between MS patients and healthy subjects, significant inverse correlations have been found between PIAS1 expression and age at disease onset. PIAS2 and PIAS3 expressions were inversely correlated with Expanded Disability Status Scale (EDSS) in patients. Moreover, PIAS3 expression was correlated with disease duration in patients and with age in controls. In addition, PIAS4 expression was inversely correlated with EDSS and age at disease onset while it was positively correlated with disease duration.
Conclusion: The present study provides evidences for altered expression of PIAS genes in IFNβ-treated MS patients compared with healthy subjects. However, future studies are needed for elaboration of their exact function in this disorder.
Keywords: multiple sclerosis, JAK/STAT pathway, protein inhibitors of activated STAT, PIAS
Corrigendum for this paper has been published
Introduction
Multiple sclerosis (MS) is a multifactorial autoimmune disorder of central nervous system.1 Among several factors implicated in this demyelinating disorder, changes in cytokines levels have crucial roles in MS pathogenesis and response to therapies.2 The Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway is a fundamental pathway for induction of cytokine functions and regulation of immune responses.3 Impairment of the JAK/STAT pathway regulation leads to imbalance between production of T helper 1 (Th1)- and Th2-type cytokines and is considered as an underlying cause of MS.2 Among transcriptional co-regulators of this pathway are protein inhibitors of activated STAT (PIAS).4 The mammalian PIAS family includes proteins encoded by four genes: PIAS1, PIAS2 (PIASx), PIAS3, and PIAS4 (PIASy).5 These proteins interact with several diverse proteins implicated in transcription and exert inhibitory effects through suppression of the DNA-binding activity of a transcription factor, enrollment of other co-regulators, or enhancing the sumoylation of a transcription factor.6 The individual interactions of PIAS1, PIAS3, PIASx, and PIASy with STAT1, STAT3, STAT4, and STAT1 have been demonstrated, respectively.7 In addition to regulation of STAT pathway through which PIAS proteins were first identified, nuclear factor-κB (NF-κB) family of transcription factors are targets of regulation by PIAS family.6 NF-κB transcription factors are extensively applied by cytokines to alter gene expression6 and their dysregulation has been regarded as a principal culprit in the pathogenesis of MS.8 Considering the role of PIAS proteins in regulation of two important pathways in modification of immune response that are altered during MS development, we aimed at expression analysis of PIAS genes in MS patients compared with healthy subjects. To the best of our knowledge, this is the first study to examine expression of PIAS genes in MS patients.
Materials and methods
Study participants
A total of 100 persons, including 48 MS patients and 52 healthy subjects participated in the current case–control study. All patients were diagnosed as relapsing–remitting MS patients based on the revised McDonald criteria.9 Patients were selected from the earlier human leukocyte antigen (HLA)-typed MS patients who were at remission due to daily injections of interferon β (IFNβ) (CinnoVex, Cinagen Company, Tehran, Iran). Based on the role of HLA-DRB1*15 as a principal risk factor for MS,10 we just included HLA-DRB1*15 negative patients in the current study. Fifty-two age- and sex-matched volunteers were enrolled in the study as control subjects. The existence of any neurological disorders was ruled out in these subjects through physical examination and history taking. The study was approved by the Ethical Committee of Shahid Beheshti University of Medical Sciences. Written informed consents were obtained from all participants. The study was conducted in accordance with the Declaration of Helsinki.
Sampling and RNA extraction
A total of 5 mL of peripheral blood was gathered from all study participants in EDTA tubes. Total RNA extraction was carried out using Hybrid-resin transfer molding blood RNA extraction Kit (Geneall Biotechnology Co., Ltd, Seoul, South Korea) from these blood samples. Subsequently, Nanodrop equipment (Thermo Scientific, Waltham, MA, USA) was applied for evaluation of RNA quantity and quality.
cDNA synthesis and quantitative reverse transcription-PCR
First strand cDNA was produced from extracted RNA samples using High-Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific) as instructed by the manufacturer. Specific primers and probes for expression analysis of PIAS1–4 genes were designed by using Allele ID 7 for x64 windows software (Premier Biosoft, Palo Alto, CA, USA). Table 1 shows the nucleotide sequence of primers and probes used in the study. HPRT1 gene was used as normalizer.
  | Table 1 Nucleotide sequence of primers and probes |
Statistical analysis
Bayesian estimation was used for evaluation of significance of difference in mean values between MS patients and healthy subjects. A normal prior distribution was supposed for parameters with 200,000 iterations. Spearman rank order correlation test was used for the evaluation of the correlation between genes expressions as well as gene expression and clinical data. The analysis of covariance (ANCOVA) was used to control the effects of possible confounding variables. P-values <0.05 were considered as significant.
Results
General information of study participants
Table 2 shows the characteristics of study participants.
  | Table 2 Characteristics of study participants Abbreviations: EDSS, Expanded Disability Status Scale; MS, multiple sclerosis; Y, years. |
Relative transcript levels of PIAS genes in MS patients and controls
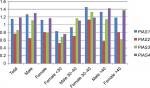
Relative expression levels of PIAS genes were not significantly different between MS patients and healthy subjects or between sex/age-based subgroups (Figure 1). Tables 3–6 show the detailed statistical parameters.
  | Table 3 PIAS1 expression in MS patients compared with healthy subjects Abbreviation: MS, multiple sclerosis. |
  | Table 4 PIAS2 expression in MS patients compared with healthy subjects Abbreviation: MS, multiple sclerosis. |
  | Table 5 PIAS3 expression in MS patients compared with healthy subjects Abbreviation: MS, multiple sclerosis. |
  | Table 6 PIAS4 expression in MS patients compared with healthy subjects Abbreviation: MS, multiple sclerosis. |
When we used ANCOVA test to control confounding effects of sex and age, we did not detect significant difference in PIAS genes expressions between MS patients and healthy subjects. Moreover, the interactions of both age and sex with disease status were insignificant.
Spearman correlation analysis showed significant pairwise correlations between expression levels of PIAS genes in all study subgroups (Spearmen correlation coefficients range: 0.4–0.88) except between PIAS3 and PIAS4 in control group. PIAS1 expression was inversely correlated with age at disease onset. PIAS2 and PIAS3 expressions were inversely correlated with Expanded Disability Status Scale (EDSS) in patients. Moreover, PIAS3 expression was correlated with disease duration in patients and with age in controls. In addition, PIAS4 expression was inversely correlated with EDSS and age at disease onset while it was positively correlated with disease duration. Table 7 shows the results of correlation analysis.
We also compared expression of PIAS genes between male and female and found no significant difference between males and females either in patient or in control groups (Table 8).
  | Table 8 Comparison of PIAS genes expression levels between male and female subjects Notes: Reference group: male, P-values computed from nonparametric frequentist methods. |
Finally, we assessed sex-specific associations between PIAS genes and clinical variables, such as EDSS and age at disease onset (Table 9). Median regression analysis showed association between PIAS3 expression and age of patients (P=0.045, 95% CI: −0.25 to –0.01). No other significant association was found after controlling the effect of sex.
  | Table 9 Results of median regression for analysis of association between PIAS genes expression and independent variables in patients Abbreviation: EDSS, Expanded Disability Status Scale. |
Discussion
Considering the role of the JAK/STAT pathway as a target of drug design in MS,3 identification of factors that modify its function is of practical value. Among regulatory mechanisms of this pathway are PIAS proteins. Notably, PIAS1 has been suggested to discriminatively decrease expression of some cytokine-responsive genes which potentiates it as a target of cytokine-based treatment modalities.6 Based on the participation of STAT and NF-kB in MS pathogenesis, we aimed at evaluation of the effects of PIAS dysregulation in MS as a prototype of autoimmune disorders. Consequently, we assessed PIAS genes expressions in IFNβ-treated MS patients and healthy subjects. We found significant correlations between the expressions of all of these genes in all blood study subgroups except for lack of correlation between PIAS3 and PIAS4 expressions in control group. This concordant expression implies the presence of a single regulatory mechanism for expression of these four genes in blood tissue or a kind of cooperative function for them. Such cooperation has been previously found between PIAS4 and PIAS1 in the regulation of the specificity and amount of NF-kB/STAT1-mediated gene stimulation.11 However, we could not find any significant difference in expression of PIAS genes between MS patients and healthy subjects. This might be due to the fact that all MS patients who participated in the current study were under treatment with IFNβ and were in remission. PIAS4 has been shown to diminish both IFN transcription and IFN-induced gene expression via various mechanisms.12 However, the effects of IFN on expression of PIAS genes have not been evaluated yet.
We have found significant inverse correlation between the expression level of PIAS1 and age at disease onset. This is in line with the previously reported role of PIAS1 in differentiation of regulatory T cells. PIAS1 has been shown to induce and retain a repressive chromatin state in the Foxp3 promoter leading to hampering the differentiation of natural regulatory T cells. Pias1 knocked-out mice have been shown to be resilient to experimental autoimmune encephalomyelitis due to the existence of high proportion of these cells.13 However, another study has shown that PIAS1 selectively decreases the expression of some STAT1- or NF-κB-dependent genes, resulting in downregulation of expression of proinflammatory cytokines and chemokines.14 Besides, we have found significant inverse correlations between EDSS and expression levels of PIAS2, PIAS3, and PIAS4. However, in spite of the observed inverse correlation between PIAS4 expression and EDSS, the expression level of this gene was inversely correlated with age at disease onset while it was positively correlated with disease duration. Such correlations might indicate certain roles of PIAS proteins in distinct phases of MS development. Moreover, it is possible that each PIAS protein exert distinctive role in the pathogenesis of MS. Future studies are needed to elaborate the effects of this gene on MS course and clinical symptoms.
In brief, in the present study, we detected significant correlations between PIAS genes expression and some clinical data of MS patients. Assessment of PIAS genes expression in a group of drug-naïve patients is necessary to elaborate the involvement of these genes in the pathogenesis of MS. Moreover, longitudinal sampling from MS patients would help to explore the effects of IFNβ on the expression of PIAS genes.
Acknowledgment
The current study was supported by a grant from Shahid Beheshti University of Medical Sciences (Grant number: 12182).
Disclosure
The authors report no conflicts of interest in this work.
References
Taheri M, Ghafouri-Fard S, Solgi G, Sayad A, Mazdeh M, Omrani MD. Determination of cytokine levels in multiple sclerosis patients and their relevance with patients’ response to Cinnovex. Cytokine. 2017;96:138–143. | ||
Hatami M, Salmani T, Arsang-Jang S, et al. STAT5a and STAT6 gene expression levels in multiple sclerosis patients. Cytokine. 2018;106:108–113. | ||
Benveniste EN, Liu Y, McFarland BC, Qin H. Involvement of the janus kinase/signal transducer and activator of transcription signaling pathway in multiple sclerosis and the animal model of experimental autoimmune encephalomyelitis. J Interferon Cytokine Res. 2014;34(8):577–588. | ||
Heinrich PC, Behrmann I, Haan S, Hermanns HM, Müller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374(Pt 1):1–20. | ||
Kim JH, Jang JW, Lee YS, et al. RUNX family members are covalently modified and regulated by PIAS1-mediated sumoylation. Oncogenesis. 2014;3:e101. | ||
Shuai K. Regulation of cytokine signaling pathways by PIAS proteins. Cell Res. 2006;16(2):196–202. | ||
Seif F, Khoshmirsafa M, Aazami H, Mohsenzadegan M, Sedighi G, Bahar M. The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun Signal. 2017;15(1):23. | ||
Mc Guire C, Prinz M, Beyaert R, van Loo G. Nuclear factor kappa B (NF-κB) in multiple sclerosis pathology. Trends Mol Med. 2013;19(10):604–613. | ||
Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69(2):292–302. | ||
Mazdeh M, Taheri M, Sayad A, et al. HLA genes as modifiers of response to IFN-β-1a therapy in relapsing-remitting multiple sclerosis. Pharmacogenomics. 2016;17(5):489–498. | ||
Tahk S, Liu B, Chernishof V, Wong KA, Wu H, Shuai K. Control of specificity and magnitude of NF-kappa B and STAT1-mediated gene activation through PIASy and PIAS1 cooperation. Proc Natl Acad Sci U S A. 2007;104(28):11643–11648. | ||
Kubota T, Matsuoka M, Xu S, et al. PIASy inhibits virus-induced and interferon-stimulated transcription through distinct mechanisms. J Biol Chem. 2011;286(10):8165–8175. | ||
Liu B, Tahk S, Yee KM, Fan G, Shuai K. The ligase PIAS1 restricts natural regulatory T cell differentiation by epigenetic repression. Science. 2010;330(6003):521–525. | ||
Liu B, Mink S, Wong KA, et al. PIAS1 selectively inhibits interferon-inducible genes and is important in innate immunity. Nat Immunol. 2004;5(9):891–898. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.