Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 14
Explore the Effect and Target of Liraglutide on Islet Function in Type 2 Diabetic Rats by miRNA Omics Technology
Authors Guo Q, Xu Y, Li J, An W, Luo D, Huang C, Huang Y
Received 22 June 2021
Accepted for publication 18 August 2021
Published 1 September 2021 Volume 2021:14 Pages 3795—3807
DOI https://doi.org/10.2147/DMSO.S325030
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Konstantinos Tziomalos
Qiuyue Guo,1 Yunsheng Xu,2 Jie Li,3 Wenrong An,3 Dan Luo,4 Chengcheng Huang,4 Yanqin Huang4
1Department of Endocrinology, Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai, 201203, People’s Republic of China; 2Department of Endocrinology, Second Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan, 250001, People’s Republic of China; 3First Clinical Medical College, Jingshi Rd. Campus, Shandong University of Traditional Chinese Medicine, Jinan, 250014, People’s Republic of China; 4Department of Endocrinology, Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan, 250014, People’s Republic of China
Correspondence: Yanqin Huang Email [email protected]; [email protected]
Purpose: To analyze the effect and potential therapeutic targets of liraglutide in type 2 diabetes through miRNA expression profiling.
Methods: Ten of 30 SPF Wistar rats, males at 4 weeks old, were randomly selected as the control group and given conventional feed, the other rats adopted high-sugar and high-fat diet combined with an intraperitoneal injection of streptozotocin to establish a T2DM model. One unsuccessful rat was excluded, and the remaining rats were randomized to the model and the liraglutide group. Liraglutide group was subcutaneously injected with liraglutide 0.11 mg/kg for 8 weeks. The biochemical indicators and staining HE were detected. The expression of miRNA in pancreatic tissue was detected by miRNA sequencing. The intersection of miRNA difference was used to predict the target gene, then functional enrichment was performed to identify its possible biological functions and signal transduction paths. Finally, qRT-PCR was used to verify the results.
Results: Compared to the model group, the level of fasting blood glucose (FBG), glucagon and insulin resistance index (HOMA-IR) in the liraglutide group were significantly decreased, fasting insulin (FINS) and insulin sensitivity index (ISI) were increased. Nine differential miRNAs (miR-135a-5p, miR-144-5p, miR-21-3p, miR-215, miR-451-5p, miR-486, miR-122-5p, miR-181d-5p and miR-345-5p) were identified at the intersection through two miRNA sequencing. A total of 3359 related target gene predictions were obtained. GO and pathway analyses demonstrated that differentially expressed genes were closely related to cell proliferation, angiogenesis, and proteolysis. Significant signaling pathways included PI signaling system, autophagy, FoxO and HIF-1 signaling pathway.
Conclusion: Liraglutide could improve islet function by regulating nine miRNAs, and the related signaling pathways included PI signaling system, autophagy, FoxO and HIF-1 signaling pathway. Our study provided the basis and direction for further exploring the molecular mechanism of liraglutide on T2DM.
Keywords: liraglutide, type 2 diabetes, miRNA expression profile, target gene, signaling pathway
Introduction
Type 2 diabetes mellitus (T2DM) is a chronic and comprehensive metabolic disease characterized by elevated blood glucose with insulin resistance and islet cell dysfunction. It is estimated that by 2045, there will be 629 million T2DM patients aged 20 and 79 years.1 As its pathogenesis is not entirely clear and its therapeutic drugs are limited, it is urgent to study the treatment of diabetes. Glucagon-like peptide-1 (GLP-1) is a type of insulinotropic that is secreted by intestinal L cells following food stimulation. After endogenous GLP-1 is degraded by dipeptidyl-peptidase -IV (DPP-IV), its biological activity is rapidly lost; its half-life in the blood is only 1–2 minutes; hence, it is difficult to be used to treat T2DM. As a novel extended-action GLP-1 receptor agonist, liraglutide overlaps 97% of the endogenous GLP-1 amino acid sequence.3 It has an amino acid substitution on the molecular structure of endogenous GLP-1, with a chain of 16 carbon fatty acids added. This structure not only prevents the degradation of liraglutide but it also extends its half-life to 13h.4 It has been proved that liraglutide regulated insulin and glucagon secretion in a glucose concentration-dependent manner, reduced blood glucose and protected islet β cells.2,5
In comparison with traditional hypoglycemic drugs, liraglutide had a variety of additional glycemic effects such as the protection of vascular endothelial cells and the reduction of the expression of inflammatory factors in diabetic kidneys.6,7 In order to further explore the anti-glycemic effects of liraglutide, this study attempted to explore its target and mechanism by microRNA (miRNA) omics technology. MiRNA is a type of single-stranded non-coding small-molecule RNA, which has nucleotides from 21 to 25, targets one or more mRNAs, and regulates gene expression through the inhibition of transcription products or degradation of target mRNAs.8 Previous studies had found that miRNAs played an important role in glucose and lipid metabolism, insulin secretion, and inflammatory response.9 MiR-29 expression attenuated insulin signal transduction, inhibited the expression of insulin receptor substrate 1 and phosphatidylinositol triokinase, and involved in regulating glucose uptake and glucose metabolism to stimulate insulin.10 MiR-24 participated in the regulation of the expression of inflammatory glycoprotein YKL-40 in cardiovascular complications of diabetes, which was closely linked to endothelial dysfunction.11
Different from the current research on liraglutide regulating a certain miRNA to treat diabetes, in this study, miRNA high-throughput sequencing was used for identifying the differential expression of pancreatic miRNAs for T2DM model rats in order to ascertain the target in liraglutide. A functional enrichment analysis was then performed to analyze its molecular mechanism, and a real-time quantitative PCR was used to verify the sequencing data. Our study explored its possible related miRNA targets and molecular mechanisms fundamentally. The corresponding findings might provide novel ideas for further research and new targets on the mechanism of liraglutide against glucose (Figure 1).
Materials and Methods
The Main Reagents
Liraglutide injection: #S20110026, Novo Nordisk, Denmark;
Streptozotocin (streptozotocin, STZ): #18883-66-4, Sigma, USA;
Sodium Citrate: #2018010203, Tianjin Zhiyuan Company;
Citric Acid: #2018-09-16, Tianjin Dingshengxin Company;
High-sugar and high-fat feed: #19093212, Beijing Keao Xieli Feed Co., Ltd.
Portable blood glucose meter: ONETOUCH-Horizon, Johnson & Johnson, USA;
Rat INS ELISA kit: #CSB-E05070r, Shanghai Keshun Biotechnology Co., Ltd.
Rat GC ELISA kit: #CSB-E12800r, Shanghai Keshun Biotechnology Co., Ltd.
GeneJET RNA Purification Kit: QIGEN, Germany;
miRcute miRNA Extraction Separation Kit: Takara, Japan;
miRcute enhanced miRNA Fluorescence Quantitative Detection Kit: Takara, Japan.
Laboratory Animals
Thirty SPF Wistar rats, male at 4-week-old, weighing (130± 10)g, were purchased from Beijing Weitong Lihua Company. The batch number was SCXK 2016-0006. They were reared in the Laboratory Animal Center of the Affiliated Hospital of Shandong University of Traditional Chinese Medicine, at a temperature of 18 ~ 22 °C, humidity 60%–70%, period of light and dark: 12h, 5 per cage, free feeding and drinking.
The animal experiments were performed according to the National Guidelines for Animal Care and Use and were approved by the Animal Ethics Committee of Shandong University of Traditional Chinese Medicine, approval number: AWE-2-19-001. The experimental procedures were in agreement with the “Regulations for the Administration of Affairs Concerning Experimental Animals“ China, for welfare of experimental animals issued by the government of the People’s Republic of China under “Laws of the People’s Republic of China”.
Establishment and Grouping of Animal Models
Ten of the 30 rats were randomly selected as a control group (n = 10) which were fed conventional food. The others were given high-sugar and high-fat diet (SPF, with a prescription of 59.5% conventional feed, 20.0% sucrose, 10.0% lard, 0.5% sodium cholate and 10.0% egg yolk powder) for 8 weeks. Then, streptozotocin (35mg/kg) was intraperitoneally injected, while the control were intraperitoneally injected with 0.1mL/100g citric acid buffer. After 72 hours, the blood was collected from the tail vein of the rats, and two consecutive measurements of fasting blood glucose (FBG) ≥ 16.7mmol/L, the T2DM model rats were successfully established. One rat with unsuccessful model was excluded, the others were randomly divided into the model (n = 9) and liraglutide groups (n = 10). The liraglutide group was given a subcutaneous injection of liraglutide at a dose of 0.11 mg/kg, while the control and model groups were given the equivalent normal saline. Each rat drank water freely. After treatment, blood was taken for 10 mL from abdominal aorta, centrifuged at 3000 rpm for 10 min, and pancreas were dissected.
Measurement Index
Determination of FBG, FINS and Glucagon (GC)
After fasting for 8–12 hours, FBG of the rats was measured by glucose oxidase. ELISA kit was adopted to detect FINS and GC, and the insulin resistance index [HOMA-IR = (FBG * FINS)/22.5] and insulin sensitivity index [ISI = (FBG * FINS) −1] were calculated.
Observation of Rat Pancreatic Tissue by Staining with Hematoxylin and Eosin
The rat pancreatic tissue was fixed with 10% formaldehyde solution for 12 hours. Paraffin-embedded after dehydration by ascending alcohol and xylene became transparent. The coagulated tissue was then cut into 4–6μm thick sections. They were HE stained and sealed after drying at 45 °C, and the morphology of the pancreas and distribution was observed. The localization of islet cells was subsequently performed under a light microscope.
miRNA Omics Technology
Total cellular RNA was extracted using TRIzol, and Gene JET RNA Purification Kit was utilized for isolation and purification. Total RNA purity was detected with Nano⁃drop, and the OD value of RNA was 260/280; 1.8–2.0;concentration≥ 500ng/µL, 28S:18S ≥1.5, RIN ≥7. After the quality of RNA was qualified, electrophoretic dephosphorylation was performed, then we added the 3ʹand 5ʹ terminal adaptor sequences for reverse transcription into cDNA. The library was constructed by PCR amplification which was then sequenced on the computer. Fastx-toolkit was used to detect raw sequencing data and perform the statistical analysis. Meanwhile, seqtk was adopted for quality control of raw data, and reference genome-specific readings were located and annotated for transcription. The screening criteria for the differential miRNAs were FC > 2 or <0.5, P < 0.05. The up-regulated and down-regulated miRNAs in the model/control group and the down-regulated and up-regulated miRNAs in the liraglutide/model group were selected respectively to obtain the differential miRNAs screened in this sequencing.
Bioinformatics Analysis
The mature sequence of miRNA was acquired from the miRBase database. The target genes of differentially expressed miRNAs were predicted by R package combined with mirwalk, targetscan, Diana MT and other miRNA target gene prediction websites, and the predicted targets of miRNAs were obtained by intersection. Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) database were used for functional annotations and enrichment pathway analyses on the predicted target genes. The selection criterion was P< 0.05.
miRNA Levels Were Verified by qRT-PCR
① Primer design: The forward primer sequences are shown in Table 1. ② miRNA was reverse transcribed into cDNA: The total RNA was extracted by mircut miRNA Isolation Kit. The total RNA’ purity and concentration were determined by enzyme-lysis instrument. Mircut enhanced miRNA cDNA first strand synthesis kit was used to reverse transcription miRNA cDNA by adding a method, which was done at 42°C 60min, 95°C 3min. cDNA was transcribed and synthesized, and stored at −20 °C; ③ Real-time PCR: PCR amplification was performed with the mircut enhanced miRNA fluorescence quantitative detection kit, the amplification of PCR according to the following procedure: 95 ° C 15min; 40 cycles (94°C 20s, 60°C 34s). Each sample was repeated three times.
 |
Table 1 Sequence Design of miRNA Primers |
The expression of miRNA in pancreas between the liraglutide group and model group was analyzed by 2−ΔΔCT method. Target miRNA primers were synthesized by Shanghai Cynes Biotechnology Co., Ltd.
Statistical Analysis
All data were statistically analyzed using SPSS version 21.0 software. The measurement data were expressed as mean ± and standard deviation (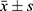 ). The comparison of two groups was carried out using the t-test. The comparison of means between multiple groups was tested by one-way analysis of variance. The difference was statistically significant (P < 0.05).
). The comparison of two groups was carried out using the t-test. The comparison of means between multiple groups was tested by one-way analysis of variance. The difference was statistically significant (P < 0.05).
Results
Effect of Liraglutide on T2DM Rats of FBG, FINS and GC
The results of FBG, FINS and GC are shown in Table 2. The FBG of three groups was in normal range prior to modeling. After modeling, the FBG in the model group was significantly higher than that in the control group. Additionally, FINS and ISI concentrations were lower than those in the control group, and GC and HOMA-IR levels were significantly increased, which were statistically significant (P < 0.05). The islet function of the model group was in a decompensated stage, and islets βcells were seriously damaged and insulin secretion was relatively insufficient.
 |
Table 2 Measurement Values of FBG, FINS, GC, ISI and HOMA-IR of Rats in Each Group (±s,n=10) |
Compared to the model group, the FBG in the liraglutide group was significantly reduced, levels of GC and HOMA-IR were relatively decreased, and levels of FINS and ISI were increased, which were statistically significant (P < 0.05). It was found that liraglutide could promote insulin secretion, improve insulin resistance, increase insulin sensitivity and effectively reduce blood glucose.
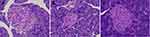
Effect of Liraglutide on Morphology of Islet Cells in T2DM Rats
In Figure 2, in the control group, pancreatic β cells were approximately round cell clusters with regular morphology that were distributed among the pancreatic acinar. The number of islet cells was numerous, the distribution was uniform, the arrangement was neat, the nuclear staining was deep, and the cytoplasm was rich.
In the model group, the islet tissue was obviously atrophic. The border was unclear and the overall shape was irregular. Moreover, the number of islet cells was significantly reduced, and their distribution was unequal, the arrangement was disordered, and some nuclear divisions were present.
In the liraglutide group, the islet morphology was found to be relatively regular and was approximately oval in shape. The number of pancreatic islets increased significantly and organized in an relatively orderly manner, with a more even distribution, deep nuclear staining, and rich cytoplasm compared to the model group (Figure 2).
Screening of Differential miRNAs and Treatment Targets Among the Three Groups
There were many significantly different miRNAs between the control and the model group, and the liraglutide group possessed many significantly different miRNAs compared with the model group, as displayed by the volcanic map in Figure 3.
Cluster analysis of these three groups of differential miRNAs was performed, which are shown in Figure 4.
According to the screening criteria (FC≥ 2 or≤ 0.5, and P < 0.05), the differential miRNA in the model group was up-regulated compared with the control group, but after liraglutide treatment, the identified inverted miRNAs included miR-135a-5p, miR-144-5p, miR-21-3p, miR-215, miR-451-5p and miR-486. The expression of miRNA in the model group was down-regulated compared with the control group, and the reversed miRNA contained miR-122-5p, miR-181d-5p, and miR-345-5p after liraglutide treatment, as described in Table 3. To a certain extent, liraglutide could regulate the expression of the nine miRNAs in pancreatic tissue of DM rats. They might be the relevant targets of liraglutide in the treatment of diabetes.
 |
Table 3 Summary of NIne miRNA Therapeutic Targets (log2FC≥1 or ≤-1, P < 0.05) |
The Regulatory Genes and Functional Analysis of the Target of miRNA
The results of the prediction demonstrated that the above nine miRNA therapeutic targets and other 3359 related genes including Tfg, Zmiz1, Neurod6, Gpr155, Meox2, Gskip, Traf2, Nlrp6, Arf4, and Srsf1 had possible target sites. GO analysis indicated that the cellular components of the targets were significantly enriched in axon part, cell front edge, myelin sheath, Golgi membrane, and cell nucleus.
The GO functional annotation showed that the cell components of nine miRNA targets were significantly enriched in the axon portion, Golgi membrane, cell leading edge, myelin sheath, and cell nucleus. Its molecular functions included cell adhesion molecule binding, ubiquitin like protease activity, transcription factor activity, phosphorus protein and so on. It was involved in the regulation of autophagy, epithelial cell proliferation, angiogenesis and other biological processes (Figure 5).
KFGG analysis indicated that the related signaling pathway involved UPP-mediated proteolysis, PI signaling system, autophagy, aldosterone synthesis and secretion, FoxO signaling pathway and HIF-1 signaling pathway (P < 0.05) (Figure 6).
Real-Time PCR Verification
Compared with miRNA sequencing, the PCR results suggested that the differential expression trend of the nine miRNAs targets was basically consistent between the model and the liraglutide group (Figure 7).
Discussion
T2DM is a metabolic disease caused by insulin resistance and islet cell dysfunction. Previous studies have found abnormal miRNA expression profile in STZ-induced diabetic rats. MiRNAs have become novel molecular markers to predict T2DM or disease progression.12 The previous study of our research group found that the traditional Chinese medicine compound Jianpi Xiaoke Decoction can also regulate the expression of miR-215, miR-135a-5p and miR-181d-5p to regulate blood sugar levels and improve the function of islet cells. Its effect might be similar to AMPK (adenosine 5‘-monophosphate (AMP)-activated protein kinase), mTOR (mammalian target of rapamycin) and other signaling pathways are related.13 In this study, miRNA high-throughput sequencing was used to screen nine differentially expressed miRNAs after the intervention of liraglutide, which were identified as follows: miR-135a-5p, miR-144-5p, miR-21-3p, miR-215, miR-451-5p, miR-486, miR-122-5p, miR-181d-5p and miR-345-5p. The corresponding differences were statistically significant. The results suggested that these miRNAs might be related to insufficiencies of islet cell function and insulin resistance. Liraglutide could regulate the expression of the aforementioned miRNAs to a certain extent and make islet function more inclined to the control group. These results indicated that liraglutide improved the function of islets regulated by these miRNAs to a certain extent.
According to literature reports, the changes in these partial differential miRNAs expression profiles were closely related to the insufficiencies of insulin secretion and the decline of islet cell function. Shao et al14 discovered that under the condition of high free fatty acids (FFAs), liraglutide could significantly promote cell proliferation and insulin secretion, and improve islet βfunction by activating PI3K/Akt and inactivating FoxO1, and the prediction of the target gene showed that miR-486 targeted the regulation of Neurod6.15 In this study, analysis of differential miRNA demonstrated that miR-486 expression was different, which was also related to neurod6, and the KEGG analysis showed that the FoxO pathway was enriched (P < 0.05). In light of the above study, it was speculated that liraglutide could regulate the blood glucose level of T2DM rats, improve the function of islet cells and promote insulin secretion, which might be related to FoxO signaling pathway and might be through activating PI3K/Akt pathway, regulating miR-486 and its downstream gene neurod6.
MiR-21 was abnormally expressed in patients with TGF and T2DM and was related to islet β-cell function and glycemic control. ZhaoH16 found that Huangqi regulated the expression of miR-144-3p and miR-486 by activating the PI3K/AKT pathway, and improve insulin resistance and change the level of serum hormone. It had been proven that miR-144, as an important marker of neuroendocrine stress response, was up-regulated in patients with T2DM and IR.17 In our study, miR-144 expression was up-regulated in the model group compared with that in the control group, and the expression was reversed after liraglutide treatment. Evidently, liraglutide was involved in regulating the expression of miR-144 and its related target genes, and miR-144 might be used as an indicator of effect of liraglutide in the treatment of T2DM. MTOR was an important protein in the insulin signalling pathway, studies showed that up-regulating the expression of miR-215 could significantly reduce the expression of mTOR, which confirmed that miR-215 was the downstream gene of mTOR.18 Based on the obtained results, liraglutide was speculated to target mTOR and bidirectionally regulate AMPK and SAD-A by regulating the expression of the miR-215 gene. In addition, some miRNAs were also observed to be involved in glucose synthesis and fat metabolism. Like miR-215, miR-135a activated the Wnt/β-catenin signaling pathway by targeting downstream genes, inhibiting the biochemical process of adipocytes and the elevated levels of triglycerides.
MiR-215 and miR-135a activated the Wnt/β-catenin signalling pathway by targeting related proteins, and inhibited the accumulation of triglycerides and the generation and differentiation of adipocytes.19–21 MiR-181b-5p participated in liver glycogen synthesis by targeting recombinant early growth response protein 1(EGR1).22
The insufficiencies of insulin secretion and the decline of islet cell function were related to islet cell apoptosis, some of the above differential miRNAs were found to play an important role in inhibiting cell proliferation and promoting apoptosis. The signalling pathway of insulin growth factor (IGF-1R) is regulated by the low expression of miR-486.23,24 Various studies have shown that miR-486-5p inhibited the proliferation of cancer cells and promotes apoptosis by inhibiting IGF-1R and its downstream mTOR, STAT3 (Signal Transducer and Activator of Transcription 3) and c-Myc.25 Additionally, miR-345-5p and miR-122 were related to apoptosis; the former regulated the proliferation, cell cycle and apoptosis of acute myeloid leukemia cells (AML) by targeting Akt2,26 while the latter inhibited cancer cell proliferation and promoted apoptosis by targeting IGF-1R (Insulin-like growth factor 1 receptor).27 The results of screening of differential genes in this experiment found that the expressions of the above miRNAs were all different (P < 0.05). In connection with the above studies, it was speculated that liraglutide could improve islet function and reduce blood glucose by regulating the expression of miR-486 in rat pancreas, which might be related to the decrease in apoptosis and the promotion of islet cell proliferation.
By analyzing the signaling pathways enriched by miRNA target genes, we found that multiple pathways were closely related to islet function and insulin secretion, such as the PI signaling system, autophagy, phosphoinositide metabolism, aldosterone synthesis and secretion, FoxO signalling pathway, and HIF-1 signalling pathway and so on. Preliminary studies had confirmed that insulin mediated the expression of miR-122 in rat liver cells through PI3K, Akt, and mTOR signaling pathways. It could be speculated that liraglutide regulated the expression of miR-122 and improved the blood glucose level of T2DM rats, which might also be related to the above pathways.28 A number of pathways interacted with glucose and lipid metabolism. MiR-451 could target glycerol kinase (Gyk), activate the Akt-FoxO1-PEPCK/G6Pase (glucose-6-phosphatase) pathway, and negatively regulate gluconeogenesis and glucose homeostasis. Overexpression of miR-451 or knockdown of Gyk could significantly inhibit liver gluconeogenesis, reduce blood glucose levels, and improve glucose tolerance.29 FoxO was the main target of insulin action and was involved in regulating gluconeogenesis, glycolysis and adipogenesis in the liver.30 Jiao Y found that adenoviruses regulated the differentiation of adipose-derived stem cells and metabolism of glucose and lipid through the PI3K/Akt/FoxO1/PPARγ (peroxisome proliferator-activated receptor γ) signalling pathway.31
In summary, our study suggested that liraglutide could improve the function of islet cells, correct the disorder of insulin secretion and regulate the level of blood glucose in rats through multiple targets and pathways.
Conclusion
In our study, nine differentially expressed miRNAs were screened for the first time by miRNA omics technology, and the potential targets of liraglutide in the treatment of T2DM were identified, and a certain number of target genes related to its regulation were predicted. These genes are strongly related to the aging and apoptosis of cells, protein metabolism and glucose transport. The related biological functions and pathways indicate that there was a potential therapeutic target exists for liraglutide in T2DM treatment. In the future, we will continue to explore the molecular mechanism of liraglutide to further reveal the mechanism of liraglutide improving islet function.
Data Sharing Statement
The original data used and analyzed in this study can be provided by the corresponding author under reasonable request.
Acknowledgments
This research was funded by the National Natural Science Foundation of China (No: 81974562, 81603613 and 81673966); Shandong Province’ Taishan Scholar Project Special Funding (NO: ts201712097) and Jinan Science and Technology Innovation Development Plan (202019029).
Author Contributions
All authors made a significant contribution to the work reported, whether that was in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; participated in drafting, revising or reviewing the article; gave final approval of the version to be published; agreed to the journal which the article is submitted; and agreed to be responsible for all aspects of the work.
Disclosure
All authors declared there was no conflict of interest in the publication of this paper.
References
1. Cho NH, Shaw JE, Karuranga S, et al. IDF diabetes atlas: global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res Clin Pract. 2018;138:271–281. doi:10.1016/j.diabres.2018.02.023
2. Renner S, Blutke A, Streckel E, Wanke R, Wolf E. Incretin actions and consequences of incretin-based therapies: lessons from complementary animal models. J Pathol. 2016;238(2):345–358. doi:10.1002/path.4655
3. Tran KL, Park YI, Pandya S, et al. Overview of glucagon-like Peptide-1 receptor agonists for the treatment of patients with Type 2 diabetes. Am Health Drug Benefits. 2017;10(4):178–188.
4. Malm-Erjefält M, Bjørnsdottir I, Vanggaard J, et al. Metabolism and excretion of the once-daily human glucagon-like peptide-1 analog Liraglutide in healthy male subjects and its in vitro degradation by dipeptidyl peptidase IV and neutral endopeptidase. Drug Metab Dispos. 2010;38(11):1944–1953. doi:10.1124/dmd.110.034066
5. Luo X, Pan L, Nie A, et al. Liraglutide protects pancreatic beta cells during an early intervention in Gato-Kakizaki rats. J Diabetes. 2013;5(4):421–428.
6. Anholm C, Kumarathurai P, Jürs A, et al. Liraglutide improves the beta-cell function without increasing insulin secretion during a mixed meal in patients, who exhibit well-controlled type 2 diabetes and coronary artery disease. Diabetol Metab Syndr. 2019;11(1):42. doi:10.1186/s13098-019-0438-6
7. Yu P, Xu X, Zhang J, et al. Liraglutide attenuates nonalcoholic fatty liver disease through adjusting lipid metabolism via SHP1/AMPK signaling pathway. Int J Endocrinol. 2019;1567095:2019. doi:10.1155/2019/1567095
8. Zhang Y, Sun X, Icli B, Feinberg MW. Emerging roles for MicroRNAs in diabetic microvascular disease: novel targets for therapy. Endocr Rev. 2017;38(2):145–168. doi:10.1210/er.2016-1122
9. Kato M, Natarajan R. MicroRNAs in diabetic nephropathy: functions, biomarkers, and therapeutic targets. Ann N Y Acad Sci. 2015;1353(1):72–88. doi:10.1111/nyas.12758
10. Massart J, Sjögren R, Lundell LS, et al. Altered miR-29 expression in Type 2 diabetes influences glucose and lipid metabolism in skeletal muscle. Diabetes. 2017;66(7):1807–1818. doi:10.2337/db17-0141
11. Deng X, Liu Y, Luo M, et al. Circulating miRNA-24 and its target YKL-40 as potential biomarkers in patients with coronary heart disease and type 2 diabetes mellitus. Oncotarget. 2017;8(38):63038–63046. doi:10.18632/oncotarget.18593
12. Yang ZM, Chen LH, Hong M, et al. Serum microRNA profiling and bioinformatics analysis of patients with type 2 diabetes mellitus in a Chinese population. Mol Med Rep. 2017;15(4):2143–2153. doi:10.3892/mmr.2017.6239
13. Guo Q, Xu Y, Li J, et al. Probe into the target and mechanism of Jianpi Xiaoke prescription for treating Type 2 diabetes mellitus through miRNA expression profiling. Evid Based Compl Alter Med. 2020;2020:7370350. doi:10.1155/2020/7370350
14. Shao S, Nie M, Chen C, et al. Protective action of Liraglutide in beta cells under lipotoxic stress via PI3K/Akt/FoxO1 pathway. J Cell Biochem. 2014;115(6):1166–1175. doi:10.1002/jcb.24763
15. Nunez Lopez YO, Garufi G, Seyhan AA. Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Mol Biosyst. 2016;13(1):106–121. doi:10.1039/c6mb00596a
16. Zhao H, Zhou D, Chen Y, Liu D, Chu S, Zhang S. Beneficial effects of Heqi san on rat model of polycystic ovary syndrome through the PI3K/AKT pathway. Daru. 2017;25(1):21. doi:10.1186/s40199-017-0188-7
17. Liang YZ, Dong J, Zhang J, Wang S, He Y, Yan YX. Identification of neuroendocrine stress response-related circulating MicroRNAs as biomarkers for Type 2 diabetes mellitus and insulin resistance. Front Endocrinol (Lausanne). 2018;9:132. doi:10.3389/fendo.2018.00132
18. Li WD, Xia JR, Lian Y. Hepatic miR‑215 target Rictor and modulation of hepatic insulin signalling in rats. Mol Med Rep. 2019;19(5):3723–3731. doi:10.3892/mmr.2019.10031
19. Tong YQ, Liu B, Zheng HY, et al. MiR-215, an activator of the CTNNBIP1/β-catenin pathway, is a marker of poor prognosis in human glioma. Oncotarget. 2015;6(28):25024–25033. doi:10.18632/oncotarget.4622
20. Li WD, Xia JR, Lian YS. Hepatic miR‑215 target Rictor and modulation of hepatic insulin signalling in rats. Mol Med Rep. 2019;19(5):3723–3731. doi:10.3892/mmr.2019.10031
21. Wei X, Cheng X, Peng Y, Zheng R, Chai J, Jiang S. STAT5a promotes the transcription of mature mmu-miR-135a in 3T3-L1 cells by binding to both miR-135a-1 and miR-135a-2 promoter elements. Int J Biochem Cell Biol. 2016;77(PtA):109–119. doi:10.1016/j.biocel.2016.06.003
22. Wang S, Liang C, Ai H, et al. Hepatic miR-181b-5p contributes to glycogen synthesis through targeting EGR1. Dig Dis Sci. 2019;64(6):1548–1559. doi:10.1007/s10620-018-5442-4
23. Peng Y, Dai Y, Hitchcock C, et al. Insulin growth factor signaling is regulated by microRNA-486, an underexpressed microRNA in lung cancer. Proc Natl Acad Sci U S A. 2013;110(37):15043–15048. doi:10.1073/pnas.1307107110
24. Ma W, Kang Y, Ning L, Tan J, Wang H, Ying Y. Identification of microRNAs involved in gefitinib resistance of non-small-cell lung cancer through the insulin-like growth factor receptor 1 signaling pathway. Exp Ther Med. 2017;14(4):2853–2862. doi:10.3892/etm.2017.4847
25. Youness RA, El-Tayebi HM, Assal RA, Hosny K, Esmat G, Abdelaziz AI. MicroRNA-486-5p enhances hepatocellular carcinoma tumor suppression through repression of IGF-1R and its downstream mTOR, STAT3 and c-Myc. Oncol Lett. 2016;12(4):2567–2573. doi:10.3892/ol.2016.4914
26. Ying X, Zhang W, Fang M, Zhang W, Wang C, Han L. miR-345-5p regulates proliferation, cell cycle, and apoptosis of acute myeloid leukemia cells by targeting AKT2. J Cell Biochem. 2018. doi:10.1002/jcb.27461
27. Wang B, Wang H, Yang Z. MiR-122 inhibits cell proliferation and tumorigenesis of breast cancer by targeting IGF1R. PLoS One. 2012;7(10):e47053. doi:10.1371/journal.pone.0047053
28. Shukla U, Tumma N, Gratsch T, et al. Abstract 5281: insulin-mediated regulation of the miR132/212 cluster and miR 122/181a expression through PI3K, Akt, mTOR signalling in primary cultured rat hepatocytes[J]. Cancer Res. 2013;73(8):5281.
29. Zhuo S, Yang M, Zhao Y, et al. MicroRNA-451 negatively regulates hepatic glucose production and glucose homeostasis by targeting glycerol kinase-mediated gluconeogenesis. Diabetes. 2016;65(11):3276–3288. doi:10.2337/db16-0166
30. Zhang W, Bu SY, Mashek MT, et al. Integrated regulation of hepatic lipid and glucose metabolism by adipose triacylglycerol lipase and FoxO proteins. Cell Rep. 2016;15(2):349–359. doi:10.1016/j.celrep.2016.03.021
31. Jiao Y, Liang X, Hou J, et al. Adenovirus type 36 regulates adipose stem cell differentiation and glucolipid metabolism through the PI3K/Akt/FoxO1/PPARγ signaling pathway. Lipids Health Dis. 2019;18(1):70.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.