Back to Journals » Drug Design, Development and Therapy » Volume 16
Exploration of the Effect and Potential Mechanism of Echinacoside Against Endometrial Cancer Based on Network Pharmacology and in vitro Experimental Verification
Authors Shu W, Wang Z, Zhao R, Shi R, Zhang J, Zhang W, Wang H
Received 23 February 2022
Accepted for publication 7 May 2022
Published 16 June 2022 Volume 2022:16 Pages 1847—1863
DOI https://doi.org/10.2147/DDDT.S361955
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tuo Deng
Wan Shu, Ziwei Wang, Rong Zhao, Rui Shi, Jun Zhang, Wei Zhang, Hongbo Wang
Department of Obstetrics and Gynecology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430030, People’s Republic of China
Correspondence: Hongbo Wang, Department of Obstetrics and Gynecology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430030, People’s Republic of China, Email [email protected]
Background: Endometrial cancer (EC) is one of the most common gynecological malignancies, especially in postmenopausal women. Echinacoside (ECH) is a major natural bioactive ingredient derived from Cistanches Herba and Echinacea that has a variety of pharmacological effects. However, the efficacy and the mechanism of ECH against EC have not been elucidated yet.
Purpose: A compound-target-disease network was constructed to explore the potential targets and mechanism of ECH against EC. Molecular docking and in vitro experiments further verified the effect of ECH against EC.
Methods: The potential targets of ECH against EC were retrieved from multiple public databases. Then, the protein–protein interaction (PPI) network was constructed to screen hub targets. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed to discover the potential mechanism. Molecular docking was utilized to verify the binding affinity between hub targets and ECH. Finally, in vitro experiments were conducted to demonstrate the anti-EC effect of ECH.
Results: A total of 110 genes were identified as potential targets of ECH against EC. The GO enrichment analysis found that targets were primarily related to oxygen species, apoptosis, and other physiological processes. KEGG pathway analysis showed that PI3K/Akt signaling pathways might play an important role in ECH against EC. Molecular docking indicated that ECH had a significant binding ability with the EGFR, AKT1, ESR1, CASP3, HSP90AA1and MMP9 targets. Results from in vitro experiments revealed that ECH induced apoptosis of Ishikawa and HEC-1-B cells by promoting the arrest of the G2M phase, increasing ROS levels, and decreasing mitochondrial membrane potential (MMP) levels. Furthermore, treatment of ECH significantly reduced the expression levels of PI3K and p-AKT, and the combination of the PI3K inhibitor (LY294002) further enhanced the effects of ECH against EC. The findings suggested that ECH exerted an inhibitory effect on EC cells by inhibiting the PI3K/AKT pathway.
Conclusion: Based on network pharmacology, molecular docking technology and in vitro experiments, we comprehensively clarified the anti-EC efficacy of ECH through multiple targets and signal pathways. Furthermore, we provided a novel idea of Traditional Chinese medicine (TCM) against EC.
Keywords: endometrial carcinoma, echinacoside, network pharmacology, mechanism, molecular docking
Introduction
Endometrial cancer (EC) is an epithelial malignancy of the endometrium that primarily affects postmenopausal women. EC is one of the three primary malignant tumors in the female genital tract, accounting for approximately 20–30% of all gynecological tumors.1 The incidence and mortality of EC are increasing every year.2 At present, surgical treatment is the primary intervention for EC. Chemotherapy and hormone therapy are the main adjuvant therapy for women with advanced and recurrent disease.3 In recent years, TCM has been applied to treat various cancers, including non-small cell lung cancer, colorectal cancer, hepatocellular cancer, and breast cancer.4–7 TCM has been shown in clinical trials to have lower toxicity and side effects than chemotherapeutic drugs. Furthermore, TCM has a specific curative effect on various diseases via various signaling pathways and targets.8,9 Recent research has revealed that natural small molecule compounds derived from TCM have the potential to be used as new anti-tumor drugs.10–12
Echinacoside (ECH) is a primary natural bioactive ingredient derived from Cistanches Herba and Echinacea that has a variety of biological effects, such as neuroprotection,13 myocardial protection,14 liver protection,15 immune regulation,16 anti-tumor17–20and other pharmacological effects. In recent years, a line of studies has indicated that ECH has an anticancer effect on various tumors.20 For example, ECH inhibits the proliferation of breast cancer cells and promotes apoptosis. Furthermore, ECH inhibits the proliferation, invasion, and migration of liver cancer cells in a dose-and time-dependent manner.19 However, no studies have been conducted to demonstrate the efficacy and mechanism of ECH against EC.
Network pharmacology is a novel method for exploring the “compound-protein/gene-disease” pathway. It visualizes the complex relationship among biological systems, drugs, and diseases by utilizing the Internet. Therefore, it provides a new perspective on the potential mechanism of drug development.7 Network pharmacology has been applied to predict protein targets from natural plant components as well as disease pathways. Molecular docking is a valuable method for designing and developing drugs based on computer-based simulations of the interaction between biological targets and drugs,21,22 It is capable of identifying active binding sites between receptors and drugs as well as determining the best binding conformation between ligands and receptors.23 In recent years, Network pharmacology and molecular docking have become valuable tools in the research on the pharmacodynamics, delineating the basis and mechanism of TCM.24,25
In this study, network pharmacology was constructed to explore the role and potential mechanism of ECH against EC, multiple databases were used to predict ECH-related targets and the effect of ECH against EC through GO and KEGG pathway enrichment analysis. Finally, the interaction between ECH and the active site of the target protein was verified by molecular docking and in vitro experiments. The workflow of the analysis procedures for ECH against EC is shown in Figure 1.
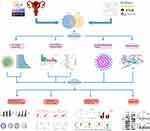 |
Figure 1 The workflow of the analysis procedures. Notes: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. |
Materials and Methods
ECH Target Gene Prediction
The structural information of ECH was retrieved from NCBI PubChem (https://pubchem.ncbi.nlm.nih.gov/). The targets of ECH were obtained by searching by five Targets fishing databases: Swiss Target Prediction Database (http://www.swisstargetprediction.ch/), Pharmmapper Database (http://www.lilab-ecust.cn/pharmmapper/), ETCM Database (http://www.nrc.ac.cn:9090/ETCM/index.PHP/home/index/), SuperPred Database (http://prediction.charite.de/) and PubChem (https://pubchem.ncbi.nlm.nih.gov/). The attributes were set to “Homo sapiens”, and the predicted targets that had normal fit scores ≥ 0.4 in PharmMapper, probability score > 0 in SWISS, known/ additionally predicted targets in SuperPred were selected as target proteins of ECH. ETCM and PubChem (https://pubchem.ncbi.nlm.nih.gov/) were used to collect targets which have been validated. Standard names for all protein targets were obtained from UniProt (http://www.uniprot.org/). The EC-related targets were obtained from three databases: GeneCards (http://www.genecards.org/), DisGeNET (http://www.disgenet.org/), and OMIM (https://omim.org/). The common targets of ECH and EC was determined by Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Protein-Protein Interaction Network Construction
The String database (https://string-db.org/) was used to analyze protein-protein interactions, The common targets were input into the STRING database to obtain interaction relationships with medium confidence basis (0.400) and “Homo sapiens”, which was saved as a TSV file. Then the analysis network was imported into Cytoscape 3.7.2 software to construct a PPI network and get the hub targets. Count.R was used to obtain the top 30 common targets of the interaction and displayed as a bar graph based on the number of nodes.
Gene Function and Pathway Analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed to analyze the common targets using the “clusterProfiler” package in R software with the P<0.05 screening condition. Annotation of gene function (Gene Ontology, GO), which includes molecular biological function (MF), biological process (BP), and cytological component (CC). The GO functions were ranked by P-value, and the top 20 pathways and top 10 GO functions were visualized as bar plots and bubble plots using the “pathview” package in R software.
Component-Target-Pathway Network Construction
To analyze the complex associations between ECH, targets and EC-related pathways, a Component-Target-Pathway (CTP) network was constructed based on Cytoscape 3.7.1 software. Cytoscape (https://cytoscape.org/download.html) is a bioinformatics software used for network analysis and construction.
Molecular Docking
Molecular docking can be used to investigate receptor-ligand interactions as well as binding patterns and affinities. In our study, molecular docking was performed to assess the interactions between ECH and key targets. The 2D structure of ECH was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), and the 3D structure of the compound was constructed by ChemOffice software. The RCSB PDB database was used to download the pdb format of the 3D structure of the core protein receptor, and PyMoL software was used to dehydrate and dephosphorylate the proteins. The Autodocktools software was then used to convert the pdb files containing the active ingredients and core protein genes to pdbqt format and search for active pockets. Finally, run AutoDock Vina for molecular binding energy calculation and molecular docking to determine the target protein’s and bioactive components’ binding affinity. AutoDock Vina was used for molecular docking and determining the binding affinity between the target protein and the bioactive ingredient, while Discovery Studio 2021 was used to align the specific docking sites between the drug molecules and the targets.
Reagents and Cell Culture
ECH (purity ≥ 98%) was purchased from Chengdu Refinside Biotechnology (Chengdu, China), The Ishikawa and HEC-1-B cell lines were purchased from Shanghai Zhongqiao Xinzhou Biotechnology Co., Ltd. Ishikawa cells were cultured in 10% fetal bovine serum and DMEM/F12 medium, and HEC-1-B cells were cultured in 10% fetal bovine serum and MEM medium. Both cell lines were cultured in a humidified atmosphere at 37 °C and 5% CO2.
CCK8 Assays
Cells in the logarithmic growth phase were inoculated at a density of 1.0×104 cells per well into a 96-well plate and cultured in a constant temperature incubator until they adhered. After treating the cells with different concentrations of ECH (200, 400, and 600 µg/mL) for 24, 36, and 48 h, respectively. The medium in each well was replaced with fresh medium containing 10% CCK8 and were incubated for 1–3 h in a constant temperature incubator. A microplate reader was used to measure the OD value at 450 nm.
Colony-Formation Assays
Cells in the logarithmic growth phase were inoculated in 6-well plates at a density of 1000 cells per well for 24 h, and after 48 h of ECH treatment, the medium was replaced with fresh medium for 14 days, with the medium changing every three days in the middle. After colony formation, cells were washed twice with PBS, fixed for 30 min with 4% paraformaldehyde, and stained with crystal violet solution (Sigma-Aldrich) (0.5% in 25% methanol). They were washed several times with PBS, air-dried, and photographed after they had dried overnight.
Apoptosis Analysis
According to the manufacturer’s instructions, EC cells were detected using the Annexin V-FITC Apoptosis Detection Kit (Beyotime Biotechnology). The adherent and suspension cells were collected, and the resuspended cells were incubated at room temperature for 15 min in the dark with Annexin V-FITC 5 µL and propidium iodide (PI) 10 µL, respectively. A flow cytometer was used to detect this (BD Biosciences). FlowJo software was used to analyze the data.
Cell Cycle Analysis
The adherent and suspension cells were collected and fixed with pre-cooled ethanol (70%) overnight at −20℃. Then incubated with propidium iodide (PI) staining solution for 30 min in the dark at room temperature and detected on the machine by flow cytometry (BD Biosciences). Data was analyzed by FlowJo software.
Measurement of the Intracellular Levels of ROS
Concentrations of intracellular ROS were measured by the ROS assay kit (Beyotime Biotechnology). The cells were incubated with dichlorofluorescein diacetate (DCFH-DA) for 30 min at 37 °C with 5% CO2. After washing the cells three times with serum-free DMEM, the intracellular levels of ROS were measured using flow cytometry (BD Biosciences).
Measurement of Mitochondrial Membrane Potential (MMP)
The mitochondrial membrane potential was measured using the JC-1 kit (Beyotime Biotechnology) following the manufacturer’s instructions. Cells were incubated with 500ul of JC-1 in a cell incubator for 20 min in the dark before being detected on the machine using flow cytometry (BD Biosciences). FlowJo software was used to analyze the data.
Western Blot Analysis
To extract total cellular protein, the cells were lysed in RIPA lysis buffer (Beyotime Institute of Biotechnology). Protein concentrations were determined using the BCA protein detection kit. Protein samples were separated using 12.5% SDS-PAGE and 10% SDS-PAGE before being transferred to PVDF membranes. The membranes were then blocked with 5% nonfat dry milk for 1 h at 37 °C before being incubated with the corresponding primary antibodies overnight at 4 °C. The membrane was then washed three times with TBST for 10 min, incubated with the secondary antibody for 2 h at 37 °C, and washed three times with TBST for ten minutes. The protein bands were visualized using an imaging system and an enhanced ECL reagent (Clinx, Shanghai, China). The results were analyzed using ImageJ software. GAPDH (ProteinTech Group, Chicago, IL, USA, 1:10000 dilution), Bcl-2 (ProteinTech Group, Chicago, IL, USA, 1:1000 dilution), Bax (ProteinTech Group, Chicago, IL, USA, 1:1000 dilution), p-AKT (ProteinTech Group, Chicago, IL, USA, 1:1000 dilution), AKT (Abcam, Cambridge, UK, 1:1000 dilution), PI3K (Abcam, Cambridge, UK, 1:1000 dilution), Caspase-3 (ProteinTech Group, Chicago, IL, USA, 1:1000 dilution) were procured.
Statistical Methods
All data in this section were statistically analyzed using the GraphPad Prism version software. Student’s t-test was used to compare statistical differences between two groups, and a one-way analysis of variance was used to compare statistical differences between three groups or more. P<0.05 was considered a significant difference.
Results
Identification of Potential Targets Against EC
A total of 396 potential ECH targets were retrieved from the Swiss Target Prediction, SuperPred, Pharmmapper, ETCM and PubChem databases (Supplementary Table S1). After removing duplicate targets, 1078 EC-related targets were collected from the GeneCards, OMIM, and DisGeNET databases. Subsequently, 110 common targets between ECH and EC genes were identified using Venny 2.1 (Supplementary Table S2), and these were considered as potential targets of ECH against EC for subsequent research (Figure 2).
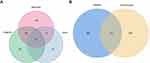 |
Figure 2 Identification of potential targets. (A) Venn diagram of EC-related targets. (B) Venn diagram of ECH targets and EC-related targets. |
Protein-Protein Interaction Network Analysis
The potential target genes of ECH were imported into the String database to determine target protein interactions and then into the Cytoscape 3.7.2 software. As shown in Figure 3A, the targets were represented as five concentric circles by the analysis network, which had 110 nodes and 1426 edges. The size and color of the nodes changed continuously with changing degree. The higher the degree, the larger the node. ALB, AKT1, EGFR, ESR1, and CASP3 were identified as hub targets to form the inner layer of concentric circles based on degree ≥70 and tight centrality ≥0.7 (Figure 3A). The corresponding bar graphs were plotted based on the node degrees to exemplify the interactions and correlations between the common targets and ECH. Further, ALB (160 edges), AKT1 (158 edges), EGFR (154 edges), ESR1 (148 edges), and CASP3 (144 edges) had a higher node degree in the top 30 hub genes, implying that they had the closest association with ECH in the treatment of EC (Figure 3B).
KEGG and GO Enrichment Analysis
To better understand the biological processes and potential mechanisms of ECH against EC, 110 targets were analyzed for GO and KEGG pathway enrichment analysis using R 4.0.3 software. The GO enrichment analysis revealed that 2051 items were related to biological process (BP), 47 items were related to cellular component (CC), and 134 items were related to molecular function (MF), indicating that the targets were primarily associated with biological process (BP). According to the corrected p-value, the biological processes mainly involved cellular response to reactive oxygen species, response to oxidative stress, cellular response to oxidative stress, response to oxidative stress reproductive system/structure development, and regulation of the apoptotic signaling pathway. The cell compositions included Ficolin-1-rich granule lumen, cytoplasmic vesicle lumen, membrane raft, membrane microdomain, and membrane region. Transmembrane receptor protein kinase/ tyrosine activity, protein serine/threonine kinase activity, phosphatase binding and endopeptidase activity were the primary molecular functions (Figure 4A). Furthermore, 149 KEGG enrichment entries were obtained, with the majority of them being enriched in cancer-related pathways such as the PI3K-Akt signaling pathway, the MAPK signaling pathway, and the Ras signaling pathway, Proteoglycans in cancer, Apoptosis, and others (Figure 4B). A bubble chart was used to screen the top 20 KEGG enrichments based on their p-values. The PI3K-Akt signaling pathway is one of the most important signaling pathways involved in the progression of EC. According to the PPI network, Akt1 is one of the hub targets, and it is directly related to the PI3K-Akt signaling pathway. As a result, we conclude that the PI3K-Akt signaling pathway may be the hub pathway of ECH against EC.
 |
Figure 4 GO and KEGG analysis. (A) GO analyses of target genes. (B) The top 20 enriched KEGG pathways of target genes. |
The Construction of Compound-Target-KEGG Network
We constructed a compound-target-KEGG pathway network based on the top 20 signaling pathways and potential targets to elucidate the mechanism of ECH against EC. As illustrated in Figure 5, the network has 132 nodes (110 genes, 1 compound, 20 KEGG pathways and 1 disease) and 597 edges, with red nodes representing EC, green nodes representing ECH, and pink oval nodes representing target nodes. The corresponding pathways are represented by green diamond nodes. The relationship between the two nodes is represented by the edges (Figure 5). The CTP network showed that ECH inhibited EC via multiple targets and pathways.
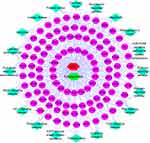 |
Figure 5 Compound-target-KEGG network, red nodes representing EC, green nodes represent ECH, and pink oval nodes represent target nodes. Green diamond nodes represent the corresponding pathways. |
Molecular Docking Analysis
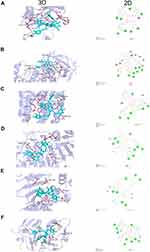
Six target genes for molecular docking with the ECH based on the KEGG pathway and degree value were selected to evaluate the compound-target binding ability at the molecular level. Binding energy was used to determine the degree of affinity between a component and its protein target. Binding energies less than −7.0 kcal/mol are thought to indicate strong ligand-receptor binding activity. Our results revealed that ECH had a high affinity for the six targets. The docking results of ECH and six target protein receptors are shown in the Figure 6. ECH and AKT1 had binding energy of −7.3 kcal/mol, primarily due to conventional hydrogen bonds. ECH could interact with ALA-58, GLN113, and TRP-80 via two hydrogen bonds in AKT1 as well as form a hydrogen bond with GLU-114, GLU113, ASP-3, SER-56, LEU-110, and GLU110 on AKT1 (Figure 6A). The binding energy of ECH and EGFR was −8.4 kcal/mol, owing primarily to hydrogen bonding, Pi-Cation, Pi-Anion, and Pi-Alkyl interaction. The structure of ECH can interact with ASP-831, THR-766, and GLU-738 via two hydrogen bonds and SER-696, LYS-721, MET-769, GLN-767, LYS-851, SER-696 via one hydrogen bond (Figure 6B). The docking binding energy of ESR1 and ECH was −8.2 kcal/mol. In ESR1, ECH could form a single hydrogen bond with GLU-323, PRO-325, GLU397, ASN-439, and ASP-321, whereas ARG-394 and TRP-393 in ESR1 formed two hydrogen bonds with ECH (Figure 6C). The binding energy of ECH and CASP3 was –9 kcal/mol. ECH could interact with ARG-164, ASP-190, GLY-125, and ASP-135 in CASP3 via one hydrogen bonds (Figure 6D). ECH had a binding energy of –8.2 kcal/mol with HSP90AA1, which had one hydrogen bond with ASP-102, GLY-132, LEU-48, SER-52, and GLY-135, two hydrogen bonds with ASN-51, and three hydrogen bonds with ASN-106 (Figure 6E). ECH bound to MMP9 with a binding energy of −8.1 kcal/mol, primarily via conventional hydrogen bonding, Pi-Pi interactions, and Pi-alkyl interactions. The ECH structure could form one hydrogen bond, two hydrogen bonds, and three hydrogen bonds with ASP-164, SER-116, and ASP-56 in MMP9 (Figure 6F), respectively. As a result of this, ECH and these protein targets exhibited strong interactions.
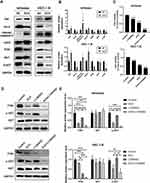
ECH Inhibits Cell Proliferation and Colony Formation in EC Cells
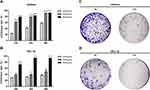
CCK8 assay was used to detect the effect of ECH on the viability of EC cells. Ishikawa and HEC-1-B cells were treated with different concentrations of ECH for 24 h, 36 h, and 48 h, respectively. The cell inhibition rate was dramatically increased in a concentration- and time-dependent manner When compared to the control group (P< 0.01) (Figure 7A and B). Colony formation experiments were further performed to confirm the anti-proliferative effect of ECH. The results showed that the number of colony formations in Ishikawa and HEC-1-B was significantly reduced after ECH treatment when compared to the control group (Figure 7C and D). These findings indicated that ECH inhibited cell proliferation and colony formation in EC cells.
ECH Promotes Apoptosis and G2M Phase Arrest in EC Cells
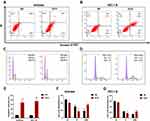
Flow cytometry was used to ensure that the anti-proliferation effect was related to apoptosis induction and cell cycle regulation. The results showed that ECH significantly increased the apoptosis rates of ISHIKAWA cells and HEC-1-B. (Figure 8A and B). The cell cycle distribution revealed that ECH induced G2/M phase arrest, which was accompanied by a decrease in G0/G1 and S phase cell numbers (Figure 8C and D). These findings demonstrated that ECH induced apoptosis in EC cells and promoted G2/M phase arrest in EC, both of which inhibited the proliferation of EC cells.
ECH Increases the Intracellular ROS Level and Reduced MMP in EC Cells
Numerous studies have demonstrated that excessive ROS production disrupts cellular homeostasis, causes oxidative stress and mitochondrial dysfunction, and induces apoptosis; thus, we investigated the effect of ECH treatment on ROS levels in EC cells. According to our findings, ECH increased the level of intracellular ROS in EC cells (Figure 9A and B). JC-1 staining was used to investigate the effect of ECH on MMP. Red JC-1 aggregates represent normal cells with high MMP, while green JC-1 monomers represent damaged cells with low membrane potential levels. A reduction in the red/green fluorescence ratio indicates a reduction in mitochondrial membrane potential, and MMP reductions were observed in both ISHIKAWA and HEC-1-B cells after 48 h of ECH treatment (Figure 9C and D). These results suggested that ECH raises ROS levels and promoted mitochondrial dysfunction.
ECH Inhibits the PI3K/AKT Pathway in EC Cells
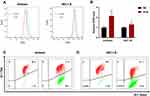
We further investigated the molecular mechanism by which ECH promotes apoptosis in EC cells. The Bcl-2 family members play an essential role in regulating apoptosis. Therefore, we investigated the expression levels of Bcl-2 and Bax in in EC cells with and without ECH treatment. Western blotting revealed that ECH increased the expression of the pro-apoptotic protein Bax while decreasing the expression of the anti-apoptotic protein Bcl-2 in Ishikawa and HEC-1-B cells. The findings suggested that the mitochondria-mediated apoptosis pathway is important in ECH-induced apoptosis, so we investigated the expression of CytoC and cleaved caspase-3 in cells to verify if ECH destroys the mitochondrial membrane stability of EC cells. ECH was discovered to increase the expression of cleaved caspase-3 and to promote the release of CytoC from mitochondria to the cytoplasm. Bioinformatics analysis identified the PI3K/Akt signaling pathway as one of the most important pathways for ECH against EC. In order to confirm that ECH inhibits EC cell proliferation by inhibiting the PI3K/AKT pathway, the effect of ECH on PI3K/AKT expression was investigated in this study. Our results showed that the PI3K/AKT pathway protein expression was significantly reduced, and the protein levels of PI3K and p-Akt were also reduced (Figure 10A and B). To further confirm whether ECH-induced apoptosis is associated with the PI3K/Akt pathway in EC cells, we measured cell viability by CCK-8 assay after treating cells with ECH, PI3K inhibitor LY294002, and ECH combined with PI3K inhibitor LY294002. The results showed that LY294002 (20 μM) enhanced the effect of ECH on cell viability (Figure 10C). The Western blot also showed that pretreatment with LY294002 further enhanced the inhibition of PI3K and p-Akt expression. It is possible to conclude that ECH has anticancer properties by inhibiting the PI3K/Akt signaling pathway (Figure 10D).
Discussion
The morbidity and mortality of EC are increasing every year with the economic development.2 There is no effective treatment for advanced and recurrent EC. TCM is the standard adjuvant therapy. In recent years, various studies have reported that TCM and its components have significant anti-tumor effects, including inhibiting tumor proliferation and migration,26 inducing apoptosis,27–29 and reversing drug resistance,30–32 among other effects. As a new way to explore drug targets, network pharmacology has become a trend in understanding the mechanism and targets of TCM.33 ECH is a small natural molecule compound with an anti-tumor effect, but its specific mechanism remains unclear. In the present study, we explored the targets and pathways of ECH on EC through network pharmacology and high-throughput molecular docking and further verification through in vitro experiments.
Network pharmacology method combined with molecular docking is a new strategy to explore the mechanism of drug action and promote the development of new drugs.7 Using this approach, we obtained 110 potential targets of ECH against EC. The PPI network showed that ALB, AKT1, EGFR, ESR1, and CASP3 may be potential targets for ECH in the treatment of EC. Estrogen receptor 1 (ESR1) is a steroid hormone receptor. Estrogen and estrogen receptor (ER) mediated signaling pathways are critical in the progression of breast cancer and EC. According to research, Erα can activate the PI3K/AKT/mTOR pathway, promoting metastasis and growth of EC.34 ESR1 activity can be reduced by estrogen signaling and control, lowering the risk of breast cancer and EC.35 The epidermal growth factor receptor (EGFR) is a tyrosine kinase that belongs to the ErbB/HER receptor tyrosine kinase family and binds to a variety of ligands. Research has demonstrated that EGFR is closely related to the prognosis of EC.36 Lapatinib (a selective EGFR and HER2 tyrosine kinase inhibitor) can inhibit the growth of EC cell lines,37 indicating that EGFR is an essential target of ECH against EC. Caspase-3 (CASP3) is a major mediator of apoptosis, which often used as a marker for cancer therapy, a line of studies has demonstrated that CASP3 is an important target for the treatment of EC.38 MMP is a metal endopeptidase that degrades the protein components of the extracellular matrix. MMP plays an important role in tumorigenesis and progression, such as tumor growth, angiogenesis, and metastasis. It has been reported that AKT1 (serine/threonine kinase 1) is a member of the AKT kinase family that is involved in cell proliferation and apoptosis. Over-expression or over-activation of Akt causes abnormal signaling. When Akt is activated on the cell membrane, it can inhibit the phosphorylation of the BAD pro-apoptotic protein, with a wide range of anti-apoptotic effects.39 The PI3K/AKT signaling pathway, which participates in cell proliferation, cell cycle, and apoptosis, is one of the most commonly dysregulated pathways in tumors. PI3K/AKT abnormal expression or over-activation has been observed in various cancers.40 The PI3K-Akt signaling pathway is a crucial pathway for cancer treatment and has been widely used in clinical trials.41 Recently, a line of studies have reported that excessive activation of the PI3K/AKT pathway plays an essential role in the occurrence and development of EC.41 Various drugs have been developed to target this pathway with the aim of reducing the growth of EC.41,42 PI3K/AKT/mTOR inhibition has shown therapeutic significance in advanced or recurrent EC.43–45 Some studies also showed that natural compounds could inhibit the progression of endometrial cancer by inhibiting PI3K/AKT signaling pathway. For example, Asparanin A from Asparagus officinalis L. induced apoptosis in EC Cells via mitochondrial and PI3K/AKT signaling pathways.46,47; The Network pharmacology results revealed that ECH induced the apoptosis of EC cells by inhibiting the PI3K/AKT1 pathway. Western blot identified that ECH significantly down-regulated the expression of p-AKT and PI3K, and LY294002 (20 μM) further increased ECH-induced cell death while enhancing the inhibition of PI3K and p-Akt expression. Therefore, ECH may have an effect on EC by inhibiting the PI3K/AKT1 pathway. We selected six molecules, including EGFR, AKT1, ESR1, CASP3, HSP90AA1 and MMP9, to further verify the network pharmacology results for molecular docking. The results indicated that ECH has a strong binding ability with these targets, supporting that ECH may play an anti-EC role by inhibiting the targets and related pathways.
Apoptosis is an essential mechanism for various drugs to exert anti-tumor effects.48,49 Exogenous and endogenous pathways are the two main in vivo pathways involved in cell apoptosis.50 The external pathway primarily causes apoptosis by inducing the binding of death receptors and ligands, which is mediated by the death receptor pathway. The mitochondrial pathways are endogenous pathways that can induce apoptosis. Excessive levels of ROS can induce mitochondrial membrane depolarization and oxidative damage to DNA, proteins, and mitochondrial lipid membranes as well as cell apoptosis.51,52 And the mitochondrial pathway is mediated by the Bcl-2 gene family, which regulates apoptosis. In the Bcl-2 family, Bax is a pro-apoptotic member, whereas Bcl-2 is an anti-apoptotic member. Bax causes mitochondrial cytochrome C release into the cytoplasm. It alters the permeability of the mitochondrial outer membrane, binds to apoptosis-related factor 1 (apaf-1),35 and then activates caspase-9 and caspase-3, resulting in a cascade of apoptotic events. Our findings showed that ECH increased intracellular ROS levels while reduced mitochondrial membrane potential levels. After ECH treatment, the anti-apoptotic gene Bcl-2 was significantly down-regulated, whereas the pro-apoptotic genes Bax, Cleaved-caspase 3, and Cyt-c were significantly up-regulated. These results elucidated that ECH induce apoptosis of EC cells through the mitochondrial pathway.
According to the first international standard for evaluating network pharmacology- “Network Pharmacology Evaluation Method Guidance”,53 we found that there are still some limitations in our work. First of all, because the resources of the public database are updated in real-time, the targets we predicted from the public network pharmacology database are limited, our study only partly clarified the molecular mechanism of ECH against EC. We still need to find more targets through more databases or further clarify the possible targets of ECH in combination with High-throughput pharmacology technology in the future. Secondly, we predicted that ECH may exert its effects through multiple targets and pathways using the network pharmacology. However, the molecular targets and pathways verified by molecular docking and experiments were limited in our study. Therefore, more experiments should be conducted to verify and explore more specific molecular mechanisms. Finally, in vivo experiments and clinical trials are required to carry out to further confirm the findings and explore the specific function of ECH against EC and lay a foundation for new drug development.
Conclusion
Through network pharmacology and molecular docking technology, we explored the efficacy of ECH against EC through multiple targets and multiple pathways. In vitro experiments further confirmed that ECH induced apoptosis of EC cells and inhibited PI3K /AKT signaling pathway. This study provides a new strategy for the treatment of EC.
Ethics and Data Sharing Statements
The OMIM, Genecards, and DisGeNET databases are publicly available and allows researchers to download and analyze. Thus, our ethics Committee of the Union Hospital of Huazhong University of Science and Technology waives ethical approval for open public databases that we used in this work.
Acknowledgments
This study was supported by grants from Major Technical Innovation Project in Hubei Province of China (2019ACA138). We greatly thank the public databases including The OMIM, Genecards, DisGeNET, Swiss Target Prediction, Pharmmapper, ETCM, SuperPred and PubChem databases for supporting this research.
Disclosure
The authors declare that there are no conflicts of interest in this work.
References
1. Brooks RA, Fleming GF, Lastra RR, et al. Current recommendations and recent progress in endometrial cancer. CA Cancer J Clin. 2019;69(4):258–279. doi:10.3322/caac.21561
2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi:10.3322/caac.21654
3. Aoki Y, Kanao H, Wang X, et al. Adjuvant treatment of endometrial cancer today. Jpn J Clin Oncol. 2020;50(7):753–765. doi:10.1093/jjco/hyaa071
4. Li Z, Feiyue Z, Gaofeng L. Traditional Chinese medicine and lung cancer–From theory to practice. Biomed Pharmacother. 2021;137:111381. doi:10.1016/j.biopha.2021.111381
5. Liao YH, Li CI, Lin CC, Lin JG, Chiang JH, Li TC. Traditional Chinese medicine as adjunctive therapy improves the long-term survival of lung cancer patients. J Cancer Res Clin Oncol. 2017;143(12):2425–2435. doi:10.1007/s00432-017-2491-6
6. Gao Y, Chen S, Sun J, et al. Traditional Chinese medicine may be further explored as candidate drugs for pancreatic cancer: a review. Phytother Res. 2021;35(2):603–628. doi:10.1002/ptr.6847
7. Zhang R, Zhu X, Bai H, Ning K. Network pharmacology databases for traditional Chinese medicine: review and assessment. Front Pharmacol. 2019;10:123. doi:10.3389/fphar.2019.00123
8. Xiang Y, Guo Z, Zhu P, Chen J, Huang Y. Traditional Chinese medicine as a cancer treatment: modern perspectives of ancient but advanced science. Cancer Med. 2019;8(5):1958–1975. doi:10.1002/cam4.2108
9. Yang Z, Zhang Q, Yu L, Zhu J, Cao Y, Gao X. The signaling pathways and targets of traditional Chinese medicine and natural medicine in triple-negative breast cancer. J Ethnopharmacol. 2021;264:113249. doi:10.1016/j.jep.2020.113249
10. Giordano A, Tommonaro G. Curcumin and cancer. Nutrients. 2019;11(10):2376. doi:10.3390/nu11102376
11. Dai X, Zhang X, Chen W, et al. Dihydroartemisinin: a potential natural anticancer drug. Int J Biol Sci. 2021;17(2):603–622. doi:10.7150/ijbs.50364
12. Yu D, Liu Y, Zhou Y, et al. Triptolide suppresses IDH1-mutated malignancy via Nrf2-driven glutathione metabolism. Proc Natl Acad Sci U S A. 2020;117(18):9964–9972. doi:10.1073/pnas.1913633117
13. Gao MR, Wang M, Jia YY, et al. Echinacoside protects dopaminergic neurons by inhibiting NLRP3/Caspase-1/IL-1β signaling pathway in MPTP-induced Parkinson’s disease model. Brain Res Bull. 2020;164:55–64. doi:10.1016/j.brainresbull.2020.08.015
14. Ni Y, Deng J, Liu X, et al. Echinacoside reverses myocardial remodeling and improves heart function via regulating SIRT1/FOXO3a/MnSOD axis in HF rats induced by isoproterenol. J Cell Mol Med. 2021;25(1):203–216. doi:10.1111/jcmm.15904
15. Tao Z, Zhang L, Wu T, Fang X, Zhao L. Echinacoside ameliorates alcohol-induced oxidative stress and hepatic steatosis by affecting SREBP1c/FASN pathway via PPARα. Food Chem Toxicol. 2021;148:111956. doi:10.1016/j.fct.2020.111956
16. Morazzoni P, Cristoni A, Di Pierro F, et al. In vitro and in vivo immune stimulating effects of a new standardized Echinacea angustifolia root extract (Polinacea). Fitoterapia. 2005;76(5):401–411. doi:10.1016/j.fitote.2005.02.001
17. Bian P, Liu C, Hu W, Ding Y, Qiu S, Li L. Echinacoside suppresses the progression of breast cancer by downregulating the expression of miR-4306 and miR-4508. Integr Cancer Ther. 2021;20:15347354211062639. doi:10.1177/15347354211062639
18. Dong L, Yu D, Wu N, et al. Echinacoside induces apoptosis in human SW480 colorectal cancer cells by induction of oxidative DNA damages. Int J Mol Sci. 2015;16(7):14655–14668. doi:10.3390/ijms160714655
19. Li W, Zhou J, Zhang Y, et al. Echinacoside exerts anti-tumor activity via the miR-503-3p/TGF-β1/Smad aixs in liver cancer. Cancer Cell Int. 2021;21(1):304. doi:10.1186/s12935-021-01890-3
20. Tang C, Gong L, Lvzi X, Qiu K, Zhang Z, Wan L. Echinacoside inhibits breast cancer cells by suppressing the Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. 2020;526(1):170–175. doi:10.1016/j.bbrc.2020.03.050
21. Pinzi L, Rastelli G. Molecular docking: shifting paradigms in drug discovery. Int J Mol Sci. 2019;20(18):18. doi:10.3390/ijms20184331
22. Ferreira LG, Dos Santos RN, Oliva G, Andricopulo AD. Molecular docking and structure-based drug design strategies. Molecules. 2015;20(7):13384–13421. doi:10.3390/molecules200713384
23. Tong H, Yu M, Fei C, et al. Bioactive constituents and the molecular mechanism of Curcumae Rhizoma in the treatment of primary dysmenorrhea based on network pharmacology and molecular docking. Phytomedicine. 2021;86:153558. doi:10.1016/j.phymed.2021.153558
24. Taha KF, Khalil M, Abubakr MS, Shawky E. Identifying cancer-related molecular targets of Nandina domestica Thunb. By network pharmacology-based analysis in combination with chemical profiling and molecular docking studies. J Ethnopharmacol. 2020;249:112413. doi:10.1016/j.jep.2019.112413
25. Li X, Lin B, Lin Z, et al. Exploration in the mechanism of fucosterol for the treatment of non-small cell lung cancer based on network pharmacology and molecular docking. Sci Rep. 2021;11(1):4901. doi:10.1038/s41598-021-84380-w
26. Gao Z, Deng G, Li Y, et al. Actinidia chinensis Planch prevents proliferation and migration of gastric cancer associated with apoptosis, ferroptosis activation and mesenchymal phenotype suppression. Biomed Pharmacother. 2020;126:110092. doi:10.1016/j.biopha.2020.110092
27. Zheng Y, Liu P, Wang N, et al. Betulinic acid suppresses breast cancer metastasis by targeting GRP78-mediated glycolysis and ER stress apoptotic pathway. Oxid Med Cell Longev. 2019;2019:8781690. doi:10.1155/2019/8781690
28. Wu CY, Yang YH, Lin YS, et al. Dihydroisotanshinone I induced ferroptosis and apoptosis of lung cancer cells. Biomed Pharmacother. 2021;139:111585. doi:10.1016/j.biopha.2021.111585
29. Li W, Li D, Kuang H, et al. Berberine increases glucose uptake and intracellular ROS levels by promoting Sirtuin 3 ubiquitination. Biomed Pharmacother. 2020;121:109563. doi:10.1016/j.biopha.2019.109563
30. Liu S, Li Q, Li G, et al. The mechanism of m(6)A methyltransferase METTL3-mediated autophagy in reversing gefitinib resistance in NSCLC cells by β-elemene. Cell Death Dis. 2020;11(11):969. doi:10.1038/s41419-020-03148-8
31. Hsu WC, Ramesh S, Shibu MA, et al. Platycodin D reverses histone deacetylase inhibitor resistance in hepatocellular carcinoma cells by repressing ERK1/2-mediated cofilin-1 phosphorylation. Phytomedicine. 2021;82:153442. doi:10.1016/j.phymed.2020.153442
32. Cheng G, Pi Z, Zhuang X, et al. The effects and mechanisms of aloe-emodin on reversing Adriamycin-induced resistance of MCF-7/ADR cells. Phytother Res. 2021;35(7):3886–3897. doi:10.1002/ptr.7096
33. Wang X, Wang ZY, Zheng JH, Li S. TCM network pharmacology: a new trend towards combining computational, experimental and clinical approaches. Chin J Nat Med. 2021;19(1):1–11. doi:10.1016/S1875-5364(21)60001-8
34. Hou X, Zhao M, Wang T, Zhang G. Upregulation of estrogen receptor mediates migration, invasion and proliferation of endometrial carcinoma cells by regulating the PI3K/AKT/mTOR pathway. Oncol Rep. 2014;31(3):1175–1182. doi:10.3892/or.2013.2944
35. Xu D, Lin TH, Yeh CR, et al. The wedelolactone derivative inhibits estrogen receptor-mediated breast, endometrial, and ovarian cancer cells growth. Biomed Res Int. 2014;2014:713263. doi:10.1155/2014/713263
36. Albitar L, Pickett G, Morgan M, Wilken JA, Maihle NJ, Leslie KK. EGFR isoforms and gene regulation in human endometrial cancer cells. Mol Cancer. 2010;9:166. doi:10.1186/1476-4598-9-166
37. Huang Y, Lin J, Yi W, et al. Research on the potential mechanism of gentiopicroside against gastric cancer based on network pharmacology. Drug Des Devel Ther. 2020;14:5109–5118. doi:10.2147/DDDT.S270757
38. Wang T, Zhang J, Hu M, et al. Differential expression patterns of glycolytic enzymes and mitochondria-dependent apoptosis in PCOS patients with endometrial hyperplasia, an early hallmark of endometrial cancer, in vivo and the impact of metformin in vitro. Int J Biol Sci. 2019;15(3):714–725. doi:10.7150/ijbs.31425
39. Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94(4):481–490. doi:10.1016/S0092-8674(00)81589-5
40. Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, González-Barón M. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30(2):193–204. doi:10.1016/j.ctrv.2003.07.007
41. Slomovitz BM, Coleman RL. The PI3K/AKT/mTOR pathway as a therapeutic target in endometrial cancer. Clin Cancer Res. 2012;18(21):5856–5864. doi:10.1158/1078-0432.CCR-12-0662
42. Chen J, Zhao KN, Li R, Shao R, Chen C. Activation of PI3K/Akt/mTOR pathway and dual inhibitors of PI3K and mTOR in endometrial cancer. Curr Med Chem. 2014;21(26):3070–3080. doi:10.2174/0929867321666140414095605
43. Roncolato F, Lindemann K, Willson ML, Martyn J, Mileshkin L. PI3K/AKT/mTOR inhibitors for advanced or recurrent endometrial cancer. Cochrane Database Syst Rev. 2019;10(10):Cd012160. doi:10.1002/14651858.CD012160.pub2
44. Myers AP. New strategies in endometrial cancer: targeting the PI3K/mTOR pathway–the devil is in the details. Clin Cancer Res. 2013;19(19):5264–5274. doi:10.1158/1078-0432.CCR-13-0615
45. Barra F, Evangelisti G, Ferro Desideri L, et al. Investigational PI3K/AKT/mTOR inhibitors in development for endometrial cancer. Expert Opin Investig Drugs. 2019;28(2):131–142. doi:10.1080/13543784.2018.1558202
46. Zhang X, Kan H, Liu Y, Ding W. Plumbagin induces Ishikawa cell cycle arrest, autophagy, and apoptosis via the PI3K/Akt signaling pathway in endometrial cancer. Food Chem Toxicol. 2021;148:111957. doi:10.1016/j.fct.2020.111957
47. Zhang F, Zhang YY, Sun YS, et al. Asparanin A from asparagus officinalis L. Induces G0/G1 cell cycle arrest and apoptosis in human endometrial carcinoma Ishikawa cells via mitochondrial and PI3K/AKT signaling pathways. J Agric Food Chem. 2020;68(1):213–224. doi:10.1021/acs.jafc.9b07103
48. Pfeffer CM, Singh ATK. Apoptosis: a target for anticancer therapy. Int J Mol Sci. 2018;19(2):448. doi:10.3390/ijms19020448
49. Hassan M, Watari H, AbuAlmaaty A, Ohba Y, Sakuragi N. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int. 2014;2014:150845. doi:10.1155/2014/150845
50. Miricescu D, Totan A, Stanescu S II, Badoiu SC, Stefani C, Greabu M. PI3K/AKT/mTOR signaling pathway in breast cancer: from molecular landscape to clinical aspects. Int J Mol Sci. 2020;22(1):173. doi:10.3390/ijms22010173
51. Yuan LQ, Wang C, Lu DF, Zhao XD, Tan LH, Chen X. Induction of apoptosis and ferroptosis by a tumor suppressing magnetic field through ROS-mediated DNA damage. Aging. 2020;12(4):3662–3681. doi:10.18632/aging.102836
52. Srinivas US, Tan BWQ, Vellayappan BA, Jeyasekharan AD. ROS and the DNA damage response in cancer. Redox Biol. 2019;25:101084. doi:10.1016/j.redox.2018.101084
53. Network Pharmacology LS. Evaluation method guidance - draft. World J Tradit Chin Med. 2021;7(1):146–154.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.