Back to Journals » OncoTargets and Therapy » Volume 14
Exosomes Derived from Pancreatic Cancer Cells Induce Osteoclast Differentiation Through the miR125a-5p/TNFRSF1B Pathway
Authors Zhou Y , Zhu Y, Dong X, Cao G, Li Y, Fan Y, Chen Q, Cai H, Wu Y
Received 6 October 2020
Accepted for publication 31 March 2021
Published 19 April 2021 Volume 2021:14 Pages 2727—2739
DOI https://doi.org/10.2147/OTT.S282319
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Gaetano Romano
Yizhao Zhou,1,2,* Yi Zhu,1,2,* Xin Dong,1,2,* Guodong Cao,1,2 Yongzhou Li,1,2 Yiqun Fan,3 Qing Chen,1,2 Haolei Cai,1,2 Yulian Wu1,2
1Department of Surgery, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, People’s Republic of China; 2Key Laboratory of Cancer Prevention and Intervention, China National Ministry of Education, Cancer Institute, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, People’s Republic of China; 3Department of Surgery, The Fourth Affiliated Hospital, Zhejiang University School of Medicine, Yiwu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yulian Wu
Department of Surgery, The Second Affiliated Hospital, Zhejiang University School of Medicine, 88 Jiefang Road, Hangzhou, 310009, People’s Republic of China
Tel/Fax +86-571-87784604
Email [email protected]
Background: Pancreatic cancer (PC) was regarded as the 4th principal cause of cancer-related fatalities in the United States and patients usually suffered from severe nutrition deficiency, muscle wasting, as well as bone loss. In our previous research, we have found that PC-derived exosomes potentially initiate insulin resistance in skeletal muscle cells. However, the role of exosomes in the PC-related bone loss remains unknown.
Methods: The effect of PC-derived exosomes on the osteoclast differentiation and femoral bone structure in the orthotopic xenograft mouse model were investigated. MiRNA expression profiles were detected and a dual luciferase experiment was conducted to identify the direct target of miRNA.
Results: Our data showed that PC-derived exosomes significantly induced osteoclast differentiation and increased expression of NFAT2, TRAP, CTSK and MMP-9. The bone volume fraction and trabecular thickness of femur significantly reduced in osteoporotic model. Microarray analyses and luciferase reporter assay showed that the process was, at least partially, mediated by the miR-125a-5p/TNFRSF1B signaling pathways.
Conclusion: According to the results, novel insights have been claimed the effect of exosomes derived from PC on bone deterioration and explained correlation between PC and cancer-related bone loss.
Keywords: pancreatic cancer, exosome, osteoclast differentiation, microRNA
Introduction
Pancreatic cancer (PC) was regarded as the 4th principal cause of cancer-related fatalities in the United States1 and was associated with poor outcomes of PC patients. In 2020, it was estimated that nearly 47,050 Americans may succumb to pancreatic cancer. Of concern, surgical resection constituted the only prospective curative therapy for PC. Nevertheless, due to either local invasion or distal metastasis, less than 20% of individuals diagnosed were identified as the patients who met the standard of surgery. The 5–year overall survival rate of PC patients was reported to be merely 9%.2,3 Notably, either chemotherapy or chemoradiation may be the effective treatments for patients unsuitable for surgery. Moreover, studies had revealed that over 80% of PC patients suffered from cachexia. The development of cachexia ultimately could causes decreased tolerance to radiotherapy and chemotherapy, lead to low overall survival.4
Cancer cachexia was a complex metabolic syndrome in over 80% of patients with cancer. Upper gastrointestinal tract cancers were characterized by relative weight loss (over 5%) or sarcopenia with weight loss (of at least 2%). For instance, anorexia, anemia, and loss of adipose and skeletal muscle mass were key clinical features of cachexia.5 Preoperative sarcopenia was associated with poor overall survival in PC patients following surgery, cachexia was suggested that may contribute to 25–30% cancer-related deaths.6–8
The underlying pathophysiology of cachexia was highly complex. There were evidences that the release of tumor cytokines may interfere with host immunity, induce paraneoplastic syndromes, leading to anorexia and hyper-catabolism.4,9 Recent studies have shown that neural infiltration presented in the muscle microenvironment were potentially associated with cancer cachexia.10 However, the intricate mechanism of cachexia was yet to be uncovered. In addition, the current clinical management of PC-related cachexia was limited and none of the available therapies can inhibit muscle wasting. An effective treatment for this disease was urgently needed.
Based on the current studies of this field, the influence of cachexia on muscle loss has been gradually investigated, whereas the bone loss and its associated molecular mechanism was still unclear. For instance, a study has revealed that bone mineral could be remarkably reduced when lung cancer patients occurred 3% weight loss.11 Similar conclusions were documented in animal model of lung, pancreatic, and colon cancer.12–14 However, the mechanism that PC affected the bone to induce cachexia and weight loss remained elusive.
Exosomes (30–150nm) were extracellular vesicles (EVs) secreted by multiple cells, as well as cancer cells. The EVs were loaded with various functional biomolecules, including DNA, mRNA, non-coding RNA (ncRNA), proteins, and bioactive lipids.15 These bioactive components can enter into recipient cells via exosomes and play a crucial role in intercellular communication.15,16 MicroRNAs (miRNAs) were single-strand endogenous non-coding RNAs (about 22 nucleotides) that target mRNAs and regulate post-transcriptional gene expression.17 Studies have shown the potential of miRNAs in regulating protein-coding genes and biological processes, including cell proliferation, differentiation, as well as apoptosis.18 EVs can protect miRNAs from being degraded by RNA enzymes and successfully transport miRNAs to recipient cells. There were increasing evidences claimed that PC cell-derived exosomes were crucial in tumor progression, metastasis, and chemoresistance19 and cause paraneoplastic β-cell dysfunction, lipolysis in adipose tissues.20,21 In our previous study, we found that pancreatic cancer cell-derived exosomes potentially initiated insulin resistance in skeletal muscle cells.22 Therefore, we hypothesized that the PC-derived exosomes can induce bone loss of cachexia through miRNA, and can be identified as potential targets for metabolic disorders.
Materials and Methods
Cell Culture
Murine pancreatic cancer cells, which were generated from the KrasLSL-G12D/+, Pdx1-Cre (KPC), as well as Trp53LSL-R172H/+ mice tumor tissues, with the potential to spontaneously grow pancreatic ductal adenocarcinoma (PDAC), were obtained from the Tingbo Liang’s Research Group (Zhejiang University, China). We kept the cell lines in RPMI-1640 growth medium (Gibco, Shanghai, China) enriched with fetal bovine serum (FBS) (20%), sodium pyruvate (100×) (1%), as well as MEM Non-Essential Amino Acids (NEAA 100×) (1%). In addition, we bought the murine pancreatic ductal epithelial cells (MPDC) from the CELLBIO Company (CBR131654, Shanghai, China). The Dulbecco’s modified Eagle’s Medium constituting L-Glutamine, sodium pyruvate (110 mg/L), as well as 4.5 g/L of D-Glucose (DMEM; Gibco, Shanghai, China) and enriched with 10% FBS was used to culture the MPDCs. We bought the RAW264.7 murine macrophage cells and 293T from ATCC, then kept in Dulbecco’s modified Eagle’s Medium consisting of L-Glutamine, sodium pyruvate (110 mg/L), as well as 4.5 g/L of D-Glucose (DMEM; Gibco, Shanghai, China) and enriched with 10% FBS.
Isolation and Identification of Exosome
Isolation of the exosomes from the supernatant of the murine pancreatic cancer cells (KPC-exosomes), as well as the murine pancreatic ductal epithelial cells (MPDC–exosomes) was performed. The cells were left to grow to a confluence of 70–80%, followed by medium replacement using a fresh medium with exosome-free FBS (processed through centrifugation for 16h at 110,000g). Following 48h of incubation, collection of the above-mentioned supernatants was conducted. After that, isolation of the exosomes was conducted through differential centrifugation as described previously.22 The exosome titers were examined using the BCA protein assay kit (Thermo, USA) after resuspension in 100ul 1× PBS. Finally, the exosomes were used for further experiments or stored at −80°C.
Zetasizer Nano Analysis
The exosome size distributions was determined through the dynamic light scattering (DLS) evaluation on a Nano Zetasizer (Malvern, UK) as per protocol provided by the manufacturer.
Electron Microscopic Viewing of Exosomes
A 20μL exosome suspension drop (1–2μg/μL) was aliquoted onto an electron-microscopy grid using a micropipette, left standing at room temperature for 3–5 minutes. Thereafter, we moved the grid to the surface of an osmium tetroxide drop (1%) (suspended in 0.1 M sodium phosphate buffer) and left to stand for 3–5 minutes prior to staining. A piece of Whatman filter paper was employed to remove the excess fluid. Lastly, the Philips Tecnai 10 transmission electron microscope (Philips, Netherlands) was employed to view the exosomes.
Exosome Internalization
Staining of the exosomes was carried out using green fluorescent probe PKH-67 (Sigma, USA) following the instructions provided by the manufacturer. After that, incubation of the PKH-67-conjugated exosomes (10g/mL) with RAW264.7 cells (25,000 cells/holes) was performed in 24-well plates for 12h. Finally, we employed a microscope (Carl Zeiss, Germany) to image the exosomes.
MTT Assay
Seeding of the RAW 264.7 cells in 96-well dish-culture plates was conducted at 3000 cells/well titer, followed by overnight incubation. Thereafter, replacement of the growth medium using complete DMEM with 0/1/5/10μg/mL of exosomes was conducted. Subsequently, 48h post-incubation, 20μL standard work solution of 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT; Sigma, USA) was introduced to the cells and further incubation for 4 h performed at 37°C. Thereafter, we completely removed the supernatant, then introduced 150μL DMSO to liquefy the violet crystals. A microplate reader (Bio-Rad, USA) was used to measure the optical density (OD).
Osteoclast Differentiation Assay
RAW264.7 cells seeded in 12-well plates were clustered into 4 groups and administered with Receptor activator of nuclear factor-κB ligand (50ng/mL) (RANKL, R&D Systems, Minneapolis) as a positive control, 10 ug/mL KPC-exosomes, 10ug/mL MPDC-exosomes or blank control. Every group had three distinct replication wells, while the assays were replicated thrice. The medium was replaced every two days. Fixation and staining of the cells was conducted using TRAP staining kit (Tartrate-resistant acid phosphatase) (Thermo, USA). For each well, 4 images were acquired. The average TRAP-positive cells/high-power field (20×) were computed.
Isolation of BMM Cells and Osteoclast Differentiation Assay
Bone marrow-derived macrophages (BMM) cells was isolated from the femurs and tibias of C57BL/6 mice (4–6 weeks of age) and cultured in a complete α-MEM medium supplemented with 10% fetal bovine serum and M-CSF-1 (30 ng/mL). Twenty-four hours later, the non-adherent cells were discarded and the adherent cells were cultured at 37°C in 5% CO2 for 3 more days. For in vitro BMM cells differentiation, BMM cells (2 × 104 cells/well) were cultured in a 24-well plate in α-MEM medium supplemented M-CSF-1 (30 ng/mL) and treated with 50ng/mL RANKL as a positive control, 50 ug/mL KPC-exosomes, 50ug/mL MPDC-exosomes or blank control. Cells were cultured for 7 days. Then, fixation and staining of the cells were conducted using TRAP staining kit (Tartrate-resistant acid phosphatase) (Thermo, USA). For each well, 4 images were acquired. The average TRAP-positive cells/high-power field (20×) were computed.
Western Blot
Lysis of the cells or exosomes was carried out using the RIPA lysis buffer (obtained from Sigma, USA) enriched with a protease inhibitor mixture (Thermo, USA). Quantification of the proteins was performed with the BCA protein assay kit (Thermo, USA). The standard protocol was followed in carrying out the assay procedures. In brief, fractionation of the proteins was done on SDS-polyacrylamide gels (10%), and the fractionated proteins were transfer-embedded onto polyvinylidene difluoride (PVDF) membranes. Thereafter, blocking of the membranes was done using 5% milk, followed by conjugation with the appropriate primary antibodies. Subsequently, the membranes were conjugated with the horseradish peroxidase (HRP)-labeled secondary antibodies prior to chemiluminescence assessment. The antibodies against C-myc, NFAT2, TSG101, as well as Alix were obtained from Abcam (USA), whereas antibodies TRAP, MMP-9 and cathepsin K(CTSK) were obtained from Santa Cruz Biotechnology (USA). Moreover, antibodies against β-actin, CD81, as well as CD9 and secondary antibodies were bought from Huabio (Hangzhou, China). Western blot densitometry was evaluated using the Image-J software (NIH, USA).
Quantitative Real-Time PCR (qRT-PCR)
The levels of gene expression were examined via qRT-PCR. Isolation of the total RNA (tRNA) from the cells or the exosomes was accomplished with the TRIzol reagent (Invitrogen, USA). After that, conversion of the tRNA to cDNA was done with the Hieff® II 1st Strand cDNA Synthesis SuperMix for qPCR Kit (Yeasen, China). The cDNA was employed in setting up the qRT-PCR reaction as the template and the Hieff® qPCR SYBR Green Master Mix Kit (Yeasen, China) utilized, with the reaction performed on a 7500 Fast™ System (Applied Biosystems, USA). GAPDH served as the internal standard. For data analysis, we used the 2−ΔΔCt approach. GenePharma (Shanghai, China) synthesized the specific miRNA primers used herein. In the miRNAs qRT-PCR, the Revert Aid First-Strand cDNA Synthesis Kit (Thermo Scientific, USA) was employed to generate the cDNAs. The small nuclear RNA (snRNA) U6 served as the internal standard.
Luciferase Reporter Assay
293T cells (2 × 104) were seeded into 24-well plates. Then, cells were transfected with pmirGLO-TNFRSF1B 3′-UTR-WT or MUT luciferase plasmid (Miaolingbil, China) and with miR-125a-5p or NC following the manufacturer’s instructions (Lipofectamine 2000, Invitrogen, USA). Luciferase assays were performed with the dual-luciferase reporter assay system (Yeasen, China) according to the manufacturer’s instructions. Luminescent signals were quantified by a luminometer (Glomax, Promega, Madison, WI, USA), and each value from the firefly luciferase construct was normalized by Renilla luciferase assay.
Orthotopic Xenograft Mouse Model
We purchased the female athymic nude mice (6-7 week-old) from the Zhejiang Chinese Medical University (Zhejiang, China) and randomly clustered into two groups (SHAM group: n = 6, KPC group: n = 8). Inoculation of the subconfluent KPC cells at a titer of 2×106 into the tail of the pancreas was performed. Mice in the SHAM group were subjected to surgical procedures, however, without injection of the tumor cells. The Animal Experimentation Committee of Zhejiang University approved the mice experiments and were carried out following the Guide to inspection of laboratory animal welfare and personnel occupational health and safety (RB/T 018-2019), as well as regulations.
Micro-CT
Right femur samples in a 16mm tube filled with PBS were a Scanco μCT40 scanner used to scan the samples at a resolution of 16µm resolution (SHAM group: n = 6, KPC group: n = 8). An experienced blinded orthopedic surgeon employed the Scanco software to reconstruct and analyze the images. Manual selection of the femoral diaphysis, as well as the distal trabeculae was performed (contoured) after every five slices, whereas morphing of the remaining was conducted to enclose the area of interest, for a total of 75 slices (1.2 mm) with the threshold at 210, as well as a gauss setting of 0. Quantification of the bone BMD, as well as the trabecular bone micro-architecture was carried out based on the mean BMD of midshaft cortical bone, and the trabecular parameters, including the bone volume fraction (bone volume over the total volume, BV/TV), trabecular number, as well as thickness.
Statistical Analysis
Data were obtained from three independent experiments and indicated as the mean ± SD. Statistical Package for the Social Sciences version 21.0 (SPSS Inc., USA) was employed for analysis. Unpaired t-tests were employed for the statistical analyses. P<0.05 (*p<0.05; **p<0.01; ***p<0.001) signified statistical significance. All histograms and curves were constructed using GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA).
Results
Morphological Profiling and Identification of Exosomes
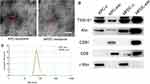
Transmission electron microscope (TEM) images showed exosomes (marked with red arrows) with different shapes (spherical or cup-shaped) (Figure 1A and B). Through particle size assessment, we demonstrated that the peak particle was approximately 70 nm and the diameter range was 30~150nm (Figure 1C), which was congruent with previous studies describing the typical morphologies of exosomes.15 Western blot analysis demonstrated typical exosome marker proteins (consisting of TSG101, Alix, CD81, and CD9) in exosomes. Also, it was found that C-myc is a nuclear protein occurring in cellular protein components bun not in the exosomal protein components (Figure 1D). Therefore, the isolated EV were exosomes.
Cancer-Derived Exosomes Readily Enter RAW 264.7 Cells
RAW 264.7 cells were incubated with 10μg/mL PKH-67-conjugated KPC-exosomes, MPDC-exosomes or KPC-exosomes without PKH-67-conjugated. The fluorescence intensity was assessed after 12 h. The majority of the green spots were aggregated in the cytoplasm of these cells, which implied that the RAW264.7 cells had the efficient potential to uptake either KPC-exosomes or MPDC-exosomes (Figure 2A).
Cancer-Derived Exosomes Promoted the Proliferation of RAW 264.7 Cells
An MTT assay was conducted to evaluate whether cancer-derived exosomes affected the viability of RAW 264.7 cells. The data revealed that treatment with 10ug/mL KPC-exosomes remarkably elevated the RAW 264.7 cell proliferation relative to the control and MPDC-exosomes (P = 0.0303 and P = 0.006, Figure 2B).
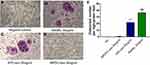
Differentiation Induction of Osteoclasts by KPC-Exosomes
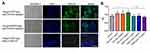
Since previous reports indicated a supportive effect of multiple myeloma-EVs on the osteoclast differentiation and activity,23,24 we assessed the influence of KPC-exosomes on the differentiation and activation of RAW264.7 cells. Indeed, KPC-exosomes significantly induced differentiation of RAW264.7 cells toward osteoclasts (marked with red arrows), where MPDC-exosomes failed with RANKL as a positive control (Figure 3A–D). Compared to MPDC-exosomes, the number of osteoclasts significantly increased in KPC-exosomes group (p<0.001, Figure 3E). Following Western blotting assays, the expression of NFAT2, TRAP, CTSK and MMP-9 in RAW264.7 cells of 10 ng/mL RANKL and 10 ul/mL KPC exosome groups were higher in different degrees as compared to the blank control and 10 ul/mL MPDC exosome groups (Figure 4A). Likewise, the mRNA expression of NFAT2, TRAP, CTSK and MMP-9 of the KPC-exosomes group were significantly higher MPDC exosome and control groups (P<0.01, Figure 4B–E).
KPC-Exosomes Induced BMM Cells Differentiate to Osteoclasts
Similarly, we isolated the BMM cells in the C57 femur and evaluated the effect of KPC-exosomes on the differentiation and activation of BMM cell. The consistent results were found. KPC-exosomes significantly induced differentiation of BMM cells toward osteoclasts (marked with red arrows), where MPDC-exosomes failed with RANKL as a positive control (Figure 5A–D). Compared to MPDC-exosomes, the number of osteoclasts significantly increased in KPC-exosomes group (p<0.001, Figure 5E).
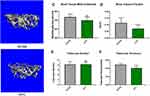
Confirmation of the Osteoporotic Model by Pancreatic Cancer
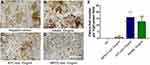
The data of the classical micro-computed tomography (Micro-CT) assessment of femoral trabecular bones are indicated in Figure 6A and B. Notably, the pancreatic tumor-carrying mice exhibited an overall reduction in trabecular bone tissue mineral density (BMD), with the mean BMD values of 735.3 ± 17.0 mgHA/cc and 698.1 ± 17.0 mgHA/cc, for the SHAM and KPC group, respectively (Figure 6C). However, the BMD of mice was lower in the KPC group than in the SHAM group but not remarkable (p=0.1049). The KPC mice showed significantly diminished Bone Volume Fraction values (p =0.0422, Figure 6D) relative to the SHAM mice, indicating loss of the bone mass due to pancreatic cancer. Though the trabecular number was not remarkably different among the groups, the bone architecture assessed by trabecular thickness was markedly lower (p=0.0038) relative to the SHAM mice (Figure 6E and F). Therefore, pancreatic cancer exhibited a remarkable decrease in bone mass.
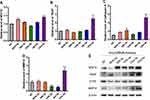
MicroRNA Microarray Analysis and Differential Screening
According to the previous miRNA chip results of our group (the specific data has been uploaded to the geodatabase, GSE95741), 1881 differentially expressed KPC-exosomal miRNAs were uncovered relative to the MPDC-exosomes (Supplementary Table S1). In order to identify prospective target of the differentially miRNA, We performed the gene prediction studies with three online analysis tools, including TargetScan, PITA and microRNA.org. As a result, a total number of 12,301 potential target were found (Supplementary Table S2). Subsequently, 281 relevant signalling pathways were classified according to KEGG functional annotations (Supplementary Table S3). Based on the screening criteria: two or at least one group (KPC-exosomes and MPDC-exosomes) have strong signals in the background of the chip (marked as Detected); Signal value difference multiples ≥2.0 and P value ≤0.05, 799 differentially expressed miRNAs were picked out, of which 339 were up-regulated, while 460 were down-regulated (Supplementary Table S4). We evaluated related signal pathways of osteoclast differentiation and identified 59 differentially expressed miRNAs (25 highly expressed and 34 lowly expressed; Figure 7A, Supplementary Table S5). To validate the microarray data, 15 miRNAs were selected from a high-expression group. Notably, RT-qPCR results showed that 5 miRNAs (miR-883b-5p, miR-666-3p, miR-125b-5p, miR-142a-3p and miR-125a-5p) were significantly highly expressed (Figure 7B).
KPC-Exosomes Up-Regulate Osteoclasts Formation via miRNA Pathways
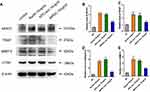
Western blot and RT-PCR tests were employed to assess the influence of these 5 miRNAs (miR-883b-5p, miR-666-3p, miR-125b-5p, miR-142a-3p and miR-125a-5p) on osteoclast activation. RT-qPCR showed that miR-125a-5p significantly elevated the expression of NFAT2, TRAP, CTSK and MMP-9 (P<0.01, Figure 8A–D). It was also found that miR-125a-5p remarkably escalated the expression of NFAT2, TRAP, CTSK and MMP-9, as demonstrated from the Western blot analysis (Figure 8E).
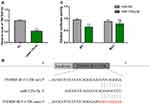
MiR-125a-5p Directly Targets TNFRSF1B
Sun et al25 reported that miR-125a-5p can significantly increase the proliferation, migration and invasion of RAW 264.7 cells and induce osteoclast differentiation. Overexpression of TNFRSF1B neutralized the promotion of cell growth and migration by miR-125a-5p, which indicated that miR-125a-5p could inhibit the expression of TNFRSF1B by binding to the 3ʹ-UTR of TNFRSF1B. To identify the relationship between miR-125a-5p and TNFRSF1B, we used qRT-PCR analysis to detect the TNFRSF1B gene in RAW 264.7 cells transfected with miR-125a-5p or miR-NC. As shown in Figure 9A, the mRNA level of TNFRSF1B was significantly inhibited by miR-125a-5p in RAW 264.7 cells. At the same time, we designed a dual fluorescein plasmid containing complementary binding the sites of TNFRSF1B with miR-125a-5p in Figure 9B. Notably, the luciferase activity of 293T cells transfected with wild type (wt) 3′-UTR of TNFRSF1B was reduced by miR-125a-5p, while the luciferase activities of 293T cells that were transfected with mutated type (mut) 3′-UTR of TNFRSF1B and miR-125a-5p or transfected with wild type (wt) 3′-UTR of TNFRSF1B and miR-NC were not affected (Figure 9C). All these results indicate that miR-125a-5p is a negative regulator of TNFRSF1B.
Discussion
Relevant studies have shown that the onset of cachexia was presented earlier about one year than confirmed PC and cachexia were early key prognostic features of PC. Cachexia can help doctors to diagnosis early stage of PC, take measures of early intervention, therefore confirmation of this syndrome earlier, always mean better prognosis.9,26 Despite it has vital clinical significance, this syndrome was still poorly diagnosed, which hindered the treatment of PC. Hence, further investigation on the mechanism of cachexia are meaningful for treatment of PC patients to improve the quality of life and prolong the overall survival.
A larger proportion of previous studies on cachexia had primarily focused on muscle loss, whereas investigations on bone loss, as well as its molecular mechanism were scant. Linton et al27 demonstrated that various PC-derived exosomes could influence macrophage polarization and function. The arachidonic acid content of the exosomes contributed to the fusogenicity of these exosomes with THP-1 macrophages and these macrophages further facilitated metastasis by producing and secreting several soluble factors. PDAC cells overexpressing certain microRNAs were reported to change their exosomal payload, which in turn could alter the macrophage polarization from M2 to M1 phenotype.28 Moreover, wild-type Panc-1 cells co-cultured with classically activated M1 macrophages can induce them to alternatively activated M2 macrophages. In our present study, we found KPC-exosomes induced differentiation of RAW264.7 cells toward osteoclasts. Micro-CT analysis demonstrated an overall reduction in trabecular bone tissue mineral density, a remarkable decrease in Bone Volume Fraction values and decreased trabecular thickness in pancreatic tumor-bearing mice (Figure 6). Besides, we found significantly increased of osteoclast-specific markers, such as NFAT2, TRAP, CTSK and MMP-9 (Figure 4) in the RAW264.7 cells treated with KPC-exosomes. Similarly, we isolated the BMM cells in the C57 femur and evaluated the effect of KPC-exosomes on the differentiation and activation of BMM cell. The consistent results were found. KPC-exosomes significantly induced differentiation of BMM cells toward osteoclasts (Figure 5). Screening investigations (microarray assessments) have revealed differentially expressed KPC-exosomal miRNAs, 59 of which were potential targets in mediating osteoclast differentiation. Previous studies have reported that miRNAs may serve pivotal roles in regulating biological processes of osteoclast differentiation and its function.29 Moreover, the upregulation of miR-21 by RANKL enhanced osteoclastogenesis though targeting programmed cell death 4 (PDCD4) protein contents, which subsequently modulated the c-Fos-NFAT2 axis.30 Based on previous reports, miR-27a alleviated adipogenesis and enhanced osteogenesis in steroid-triggered rat bone marrow mesenchymal stem cells (BMSCs) through targeting peroxisome proliferator-activated receptor gamma (PPARγ), as well as gremlin 1 (GREM1).31 Despite the recognized importance of miRNAs in the modulation of bone remodeling, limited information has been published to deeply claim the precise role and mechanism of PC-derived exosomes in osteoclastogenesis.
Herein, we detected the differently expression of miRNAs between KPC-exosomes and MPDC-exosomes and found 25 up-regulated miRNA related with signal pathways of osteoclast differentiation (Figure 7A). To validate the microarray data, 15 miRNAs were selected from a high-expression group and RT-qPCR results showed that 5 miRNAs (miR-883b-5p, miR-666-3p, miR-125b-5p, miR-142a-3p and miR-125a-5p) were significantly highly expressed (Figure 7B). RT-qPCR and Western blot analysis showed that miR-125a-5p significantly elevated the expression of NFAT2, TRAP, CTSK and MMP-9, which are the marker gene of osteoclast (Figure 8). So, the miR-125a-5p may play an important role in osteoclast differentiation. Notably, MiR-125a-5p was extensively expressed in distinct tissues, as well as cells, it targets distinct genes to modulate the occurrence of cancer,32,33 skeletal muscle cell differentiation,34 immunity,35 diagnosis of heart failure,36 and induces pulmonary arterial hypertension.37 Sun et al25 suggested miR-125a-5p could remarkably increase the in vitro proliferation, migration, as well as infiltration of RAW 264.7 cells and induced osteoclast differentiation. The enhancing influence of miR-125a-5p on the growth, as well as mobility of cells was neutralized by over-expression of TNFRSF1B. To identify the relationship between miR-125a-5p and TNFRSF1B, we found the mRNA level of TNFRSF1B was significantly inhibited by miR-125a-5p in RAW 264.7 cells (Figure 9A). At the same time, we performed a Luciferase reporter assay and the luciferase activity of 293T cells transfected with wild type (wt) 3′-UTR of TNFRSF1B was reduced by miR-125a-5p (Figure 9B and C). All the consistent results indicate that miR-125a-5p is a negative regulator of TNFRSF1B.
In conclusion, we established that PC-derived exosomes potentially induce osteoclast differentiation and the process was at least partially mediated by the miR-125a-5p/TNFRSF1B signaling pathways. We provided a novel insight into the mechanism of the effect of exosomes derived from PC on bone deterioration, these may help us to deal with the correlation between PC and cancer-related bone loss. In brief, this study have provided us a new strategy to know and treat cancer cachexia, that may help us to improve the diagnosis and treatment of PC. Further studies need to be done to recommend more promising PC-derived-exosomal-miRNAs, and confirm the mechanism.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China: Nos. 81772562 and 81572922 (Dr. Yulian Wu), the National Youth Science Foundation of China: No.81702316 (Dr. Yi Zhu) and Project of science and technology of traditional Chinese medicine of Zhejiang Province: No. 2017ZA088 (Dr. Haolei Cai).
Disclosure
The authors reported no conflicts of interest for this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
2. Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395(10242):2008–2020. doi:10.1016/S0140-6736(20)30974-0
3. Ilic M, Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol. 2016;22(44):9694–9705. doi:10.3748/wjg.v22.i44.9694
4. Baracos VE, Martin L, Korc M, Guttridge DC, Fearon KCH. Cancer-associated cachexia. Nat Rev Dis Primers. 2018;4(1):17105. doi:10.1038/nrdp.2017.105
5. Fearon K, Strasser F, Anker SD, et al. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol. 2011;12(5):489–495. doi:10.1016/S1470-2045(10)70218-7
6. Peng P, Hyder O, Firoozmand A, et al. Impact of sarcopenia on outcomes following resection of pancreatic adenocarcinoma. J Gastrointest Surg. 2012;16(8):1478–1486. doi:10.1007/s11605-012-1923-5
7. Sandini M, Bernasconi DP, Fior D, et al. A high visceral adipose tissue-to-skeletal muscle ratio as a determinant of major complications after pancreatoduodenectomy for cancer. Nutrition. 2016;32(11–12):1231–1237. doi:10.1016/j.nut.2016.04.002
8. von Haehling S, Anker MS, Anker SD. Prevalence and clinical impact of cachexia in chronic illness in Europe, USA, and Japan: facts and numbers update 2016. J Cachexia Sarcopenia Muscle. 2016;7(5):507–509. doi:10.1002/jcsm.12167
9. Sah RP, Nagpal SJ, Mukhopadhyay D, Chari ST. New insights into pancreatic cancer-induced paraneoplastic diabetes. Nat Rev Gastroenterol Hepatol. 2013;10(7):423–433. doi:10.1038/nrgastro.2013.49
10. Imoto A, Mitsunaga S, Inagaki M, et al. Neural invasion induces cachexia via astrocytic activation of neural route in pancreatic cancer. Int J Cancer. 2012;131(12):2795–2807. doi:10.1002/ijc.27594
11. Fearon KC. The Sir David Cuthbertson Medal lecture 1991. The mechanisms and treatment of weight loss in cancer. Proc Nutr Soc. 1992;51(2):251–265. doi:10.1079/PNS19920036
12. Choi E, Carruthers K, Zhang L, et al. Concurrent muscle and bone deterioration in a murine model of cancer cachexia. Physiol Rep. 2013;1(6):e00144. doi:10.1002/phy2.144
13. Zhang Q, Sun X, Yang J, et al. ZIP4 silencing improves bone loss in pancreatic cancer. Oncotarget. 2015;6(28):26041–26051. doi:10.18632/oncotarget.4667
14. Bonetto A, Kays JK, Parker VA, et al. Differential bone loss in mouse models of colon cancer cachexia. Front Physiol. 2016;7:679.
15. Tkach M, Thery C. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164(6):1226–1232. doi:10.1016/j.cell.2016.01.043
16. Milane L, Singh A, Mattheolabakis G, Suresh M, Amiji MM. Exosome mediated communication within the tumor microenvironment. J Control Release. 2015;219:278–294. doi:10.1016/j.jconrel.2015.06.029
17. Tesfaye AA, Azmi AS, Philip PA. miRNA and gene expression in pancreatic ductal adenocarcinoma. Am J Pathol. 2019;189(1):58–70. doi:10.1016/j.ajpath.2018.10.005
18. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi:10.1038/nrg3074
19. Lan B, Zeng S, Grutzmann R, Pilarsky C. The role of exosomes in pancreatic cancer. Int J Mol Sci. 2019;20(18):4332. doi:10.3390/ijms20184332
20. Javeed N, Sagar G, Dutta SK, et al. Pancreatic cancer-derived exosomes cause paraneoplastic beta-cell dysfunction. Clin Cancer Res. 2015;21(7):1722–1733. doi:10.1158/1078-0432.CCR-14-2022
21. Sagar G, Sah RP, Javeed N, et al. Pathogenesis of pancreatic cancer exosome-induced lipolysis in adipose tissue. Gut. 2016;65(7):1165–1174. doi:10.1136/gutjnl-2014-308350
22. Wang L, Zhang B, Zheng W, et al. Exosomes derived from pancreatic cancer cells induce insulin resistance in C2C12 myotube cells through the PI3K/Akt/FoxO1 pathway. Sci Rep. 2017;7(1):5384. doi:10.1038/s41598-017-05541-4
23. Faict S, Muller J, De Veirman K, et al. Exosomes play a role in multiple myeloma bone disease and tumor development by targeting osteoclasts and osteoblasts. Blood Cancer J. 2018;8(11):105. doi:10.1038/s41408-018-0139-7
24. Raimondi L, De Luca A, Amodio N, et al. Involvement of multiple myeloma cell-derived exosomes in osteoclast differentiation. Oncotarget. 2015;6(15):13772–13789. doi:10.18632/oncotarget.3830
25. Sun L, Lian JX, Meng S. MiR-125a-5p promotes osteoclastogenesis by targeting TNFRSF1B. Cell Mol Biol Lett. 2019;24(1):23. doi:10.1186/s11658-019-0146-0
26. Singhi AD, Koay EJ, Chari ST, Maitra A. Early detection of pancreatic cancer: opportunities and challenges. Gastroenterology. 2019;156(7):2024–2040. doi:10.1053/j.gastro.2019.01.259
27. Linton SS, Abraham T, Liao J, et al. Tumor-promoting effects of pancreatic cancer cell exosomes on THP-1-derived macrophages. PLoS One. 2018;13(11):e0206759. doi:10.1371/journal.pone.0206759
28. Su MJ, Aldawsari H, Amiji M. Pancreatic cancer cell exosome-mediated macrophage reprogramming and the role of micro-RNAs 155 and 125b2 transfection using nanoparticle delivery systems. Sci Rep. 2016;6(1):30110. doi:10.1038/srep30110
29. Inoue K, Nakano S, Zhao B. Osteoclastic microRNAs and their translational potential in skeletal diseases. Semin Immunopathol. 2019;41(5):573–582. doi:10.1007/s00281-019-00761-4
30. Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood. 2011;117(13):3648–3657. doi:10.1182/blood-2010-10-311415
31. Gu C, Xu Y, Zhang S, et al. miR-27a attenuates adipogenesis and promotes osteogenesis in steroid-induced rat BMSCs by targeting PPARgamma and GREM1. Sci Rep. 2016;6(1):38491. doi:10.1038/srep38491
32. Kim JK, Noh JH, Jung KH, et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57(3):1055–1067. doi:10.1002/hep.26101
33. Nishida N, Mimori K, Fabbri M, et al. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 2011;17(9):2725–2733. doi:10.1158/1078-0432.CCR-10-2132
34. Li R, Li B, Shen M, et al. LncRNA 2310043L19Rik inhibits differentiation and promotes proliferation of myoblast by sponging miR-125a-5p. Aging (Albany NY). 2020;12(7):5625–5639. doi:10.18632/aging.102905
35. Essandoh K, Li Y, Huo J, Fan GC. MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock. 2016;46(2):122–131. doi:10.1097/SHK.0000000000000604
36. Wong LL, Armugam A, Sepramaniam S, et al. Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur J Heart Fail. 2015;17(4):393–404. doi:10.1002/ejhf.223
37. Cai Z, Li J, Zhuang Q, et al. MiR-125a-5p ameliorates monocrotaline-induced pulmonary arterial hypertension by targeting the TGF-beta1 and IL-6/STAT3 signaling pathways. Exp Mol Med. 2018;50(4):45. doi:10.1038/s12276-018-0068-3
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.