Back to Journals » OncoTargets and Therapy » Volume 17
Exosome-Delivered EGFR Induced by Acidic Bile Salts Regulates Macrophage M2 Polarization to Promote Esophageal Adenocarcinoma Cell Proliferation
Authors Chen C, Ding J , Ma Z, Xie Y, Zhang L, Zhu D
Received 28 August 2023
Accepted for publication 5 February 2024
Published 16 February 2024 Volume 2024:17 Pages 113—128
DOI https://doi.org/10.2147/OTT.S437560
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Chuangui Chen,1,2,* Jinsheng Ding,3,* Zhao Ma,1 Yongjie Xie,3 Linhua Zhang,4 Dunwan Zhu4
1Department of Minimally Invasive Esophagus Surgery, Tianjin’s Clinical Research Center for Cancer, National Clinical Research Center of Cancer, Tianjin Medical University Cancer Institute and Hospital, Key Laboratory of Cancer Prevention and Therapy, Tianjin, 300060, People’s Republic of China; 2Beijing Viewsolid Biotechnology Co., LTD, Beijing, 102200, People’s Republic of China; 3Department of Pancreatic Cancer, Tianjin’s Clinical Research Center for Cancer, National Clinical Research Center of Cancer, Tianjin Medical University Cancer Institute and Hospital, Key Laboratory of Cancer Prevention and Therapy, Tianjin, 300060, People’s Republic of China; 4Tianjin Key Laboratory of Biomedical Materials, Key Laboratory of Biomaterials and Nanotechnology for Cancer Immunotherapy, Institute of Biomedical Engineering, Chinese Academy of Medical Sciences & Peking Union Medical College, Tianjin, 300192, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Dunwan Zhu, Tianjin Key Laboratory of Biomedical Materials, Key Laboratory of Biomaterials and Nanotechnology for Cancer Immunotherapy, Institute of Biomedical Engineering, Chinese Academy of Medical Sciences & Peking Union Medical College, 236 Baidi Road, Nankai District, Tianjin, 300192, People’s Republic of China, Tel +86-13034380016, Fax +86-2223340123, Email [email protected]
Purpose: Chronic gastroesophageal reflux disease (GERD) causes the abnormal reflux of acid and bile salts, which would induce Barrett’s esophagus (BE) and esophageal adenocarcinoma (EAC). EGFR, as one of main components of the exosome, plays an important role in cancer progression. Here, we investigated the role of acidic bile salts (ABS)-induced exosomal EGFR in EAC cell proliferation.
Methods: Electronic microscopic examination and Western blot were used to identify exosomes. Western blot, siRNA transfection, enzyme-linked immunosorbent assay, qRT-PCR, cell viability detection, mouse xenograft tumor models, and immunohistochemical staining were performed to study the function of ABS-induced exosomal EGFR in cell proliferation.
Results: We found that ABS improved the exosomal EGFR level of normal human esophageal epithelial cells, BE cells, and BE-associated adenocarcinoma cells. The results were confirmed in the serum-derived exosomes from healthy persons and patients suffering from GERD, BE with or without GERD, and EAC with or without GERD. Moreover, cell line-derived exosomal EGFR was found to promote macrophage M2 polarization through the PI3K-AKT pathway. The co-incubation medium of macrophages and exosomes improved cell proliferation and tumor growth, which depended on the exosomal EGFR level. CCL18 was identified as the most effective component of the co-incubation medium to promote EAC cell proliferation by binding to its receptor PITPNM3 in vitro and in vivo.
Conclusion: Our findings demonstrate that ABS-induced exosomal EGFR regulates macrophage M2 polarization to promote EAC proliferation. This study provides an important insight into the role of ABS in EAC development.
Keywords: cancer progression, pro-oncogenic, PI3K/AKT pathway, CCL18, PITPNM3
Introduction
Esophageal cancer is highly lethal, leading to more than half a million deaths yearly. It has two main histological subtypes: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). Although EAC only accounts for 20% of EC cases, its survival rate is very low. The incidence has increased rapidly in the past 40 years.1 Barrett’s esophagus (BE) is one of the main risk factors for EAC, characterized by columnar metaplasia in the distal esophagus, and is a well-established precancerous state of EAC.2 As one of the main risk factors of BE, chronic gastroesophageal reflux disease (GERD) causes the abnormal reflux of acid and bile salts into the esophagus, which can regulate oncogenic signaling pathways to promote the progression of BE and EAC.3–5 Nevertheless, the detailed underlying mechanism is still elusive.
Exosomes (~100 nm on average), a subset of extracellular vesicles, are secreted by cells and mediate signaling transduction between neighboring or distant cells. Exosomes contain many cellular constituents, including membrane proteins, cytosolic and nuclear proteins, metabolites, and nucleic acids.6 There is emerging evidence that cancer-derived exosomes are closely related to the tumor microenvironment (TME). The exosomal components can be delivered to recipient cells to regulate immune response and cancer progression.7,8 Epidermal growth factor receptor (EGFR), as a membrane protein, is one of the main components of the exosome. It is a potential cancer biomarker9 and is well known to play a dominant role in the pathogenesis and development of various cancers.10 For instance, gastric cancer cells can secret exosomes containing EGFR, which can be delivered into the liver stromal cells and regulate the liver microenvironment to promote metastatic cancer cells landing and proliferation by activating hepatocyte growth factor.11 Human papillomavirus can enhance the expression of EGFR in exosomes, which is involved in the epithelial-mesenchymal transition (EMT) of lung cancer cells.12 In lung cancer, EGFR-containing exosomes can also regulate the function of T cells.13 Moreover, EGFR in breast cancer cell-derived exosomes can activate the mitogen-activated protein kinase (MAPK) survival pathway and stimulate monocyte survival.14 However, the role of EGFR-containing exosomes from EAC is still unclear.
Within the TME, the most prominent immune cell is tumor-associated macrophages (TAMs). TAMs may produce a variety of cytokines to influence cancer development and progression.15 Macrophages are divided into M1 and M2 phenotypes that define the possible extremes of in vitro- or in vivo-polarized cells.16 The two phenotypes can be induced to transform each other under different conditions and have opposite functions in tumor growth and metastasis.M1-TAMs produce antitumor immunity-related factors (eg, IL-1, IL-12, and CXCL10) to inhibit tumor proliferation, invasion, metastasis, and angiogenesis; however, M2-TAMs secrete tumor promotion-related factors (eg, IL-10 and VEGF) to play a prompting role in tumor progression.17,18 In breast cancer cells, glycyrrhetinic acids were found to strongly suppress M2 macrophage-induced cell proliferation and migration, which was mediated by the JNK signaling pathway.19 M2 macrophages can lead to the accumulation of leukemia blast cells with low calreticulin.20 Compared to other cancers, studies on esophageal cancer are very limited. A meta-analysis showed M2 macrophage infiltration was associated with poor overall survival of ESCC.21 M2 macrophage infiltration following M1 macrophage infiltration can promote the development of esophageal cancer.22 In one study, EAC cells were found to polarize THP-1 cells into M2 macrophages, promoting EAC cell migration and invasion.23 However, the functions of TAMs, especially macrophage polarization, in EAC progression have been largely unknown.
This study found that acidic bile salts (ABS) upregulate exosomal EGFR levels of normal human esophageal cells, BE, and EAC cells. We further investigated the roles of exosome-delivered EGFR inTAMs polarization and EAC cell proliferation in vitro and in vivo. Our results provide a new perspective regarding the molecular mechanism for acid and bile salts’ role in EAC occurrence and progression.
Materials and Methods
Cells Culture
Normal human esophageal epithelial cell line HEEC was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, PR China), BE cell line CPB was obtained from American Type Culture Collection, andBE-associated adenocarcinoma cell line OE33 were obtained from Sigma-Aldrich (St. Louis, MO, USA). OE33 cell lines stably expressing EGFR shRNA, PITPNM3 shRNA, or scrambled shRNA were purchased from ViewSolid Biotech (Beijing, China). Cells were cultured using RPMI 1640 (Invitrogen, USA) with 10% exosomes-free fetal bovine serum (Gibco, USA), streptomycin (100 μg/mL), and penicillin (100 U/mL) at 37°C with 5% CO2.
Serum Samples, Reagents, and Antibodies
From September 2021 to June 2022, serum samples were collected from healthy adults, patients suffering from BE with or without GERD, and patients suffering from EAC with or without GERD (n=25) in Tianjin Medical University Cancer Institute and Hospital. All patients with GERD were identified to have reflux of gastric acid and bile. All serum donors signed informed consent. Our study complies with the Declaration of Helsinki. The samples were immediately processed for subsequent exosomes isolation. The ABS cocktail consists of an equimolar mixture of sodium salts of glycocholic acid, taurocholic acid, glycodeoxycholic acid, deoxycholic acid (CALBIOCHEM, La Jolla, CA, USA), and glycochenodeoxycholic acid (Sigma-Aldrich, Germany), which was used to mimic the mixture of bile acids in the distal esophagus during GERD.24 We used a final concentration of 100 µM of the ABS cocktail in all experiments. LY294002 (an inhibitor of the PI3K/AKT signaling pathway) was obtained from Abcam (Britain). CCL18recombinant protein (MBS553104) was purchased from MyBioSource (USA). Antibodies used in this study are shown in Table 1.
 |
Table 1 Discription of Antibodies Used in the Study |
Exosomes Isolation and Identification
HEEC, CPB or OE33 cells were cultured for 48 h. Cell culture media were collected by centrifuging at 5000rpm for 10 min. Serum was extracted by centrifugation of peripheral blood samples at 4000 rpm (4 °C for 10 min) and then at 12,000 rpm (4 °C for 20 min). Exosomes were isolated using Exosome Purification Kit (57600, NorgenBiotek, Canada) according to the instructions. Briefly, cell culture media or serum was added with a series of reagents and centrifuged, and then the final obtained supernatant was transferred to a filter spin column to obtain the purified exosomes. The vesicles were identified by transmission electron microscopic examination and exosome markers (TSG101, Annexin, and CD9). The concentrations of exosomes were determined by Zetaview (Particle Metrix, Germany).
Preparation of Co-Culture Medium
Human THP-1 monocytes were grown in six-well plates and treated with 100 ng/mL phorbol myristate acetate (PMA) for 24h to differentiate into adherent M0 macrophages. M0 macrophages were incubated with different cell-derived exosomes (25 μg/mL) for 24 h. The co-culture medium was collected by centrifuging at 10,000 g for 5 min and then partitioned and stored at −20°C for use.
Western Blot
Total protein from cells or exosomes was extracted with RIPA Lysis Buffer (Beyotime, Shanghai, China) and quantified via bicinchoninic acid (BCA) (Pierce, Waltham, MA, USA) way. A certain amount of protein was loaded for electrophoresis, and then the protein was shifted to a PVDF (Beyotime, Shanghai, China). The membranes were soaked in 5% skim milk for about 2h, and were immersed in primary antibodies at 4°C overnight and then were incubated with secondary antibodies for 2–3h at room temperature. Protein bands were visualized by enhanced chemiluminescence (ECL) and calculated by ImageJ2× Software.
siRNA Transfection
siRNA oligonucleotides were synthesized by ViewSolid Biotech (Beijing, China) and were transfected into cells according to the instruction of Lipofectamine 3000 (Invitrogen, USA). siRNA sequences are shown in the following: EGFR siRNA sequence: EGFR siRNA sequence: 5’-UAAUUCCUCUGCACAUAGGUA-3’. PITPNM3 siRNA sequence:5’- CGCGCAUGAUCCUGCGCAATT-3’. Scramble siRNA sequence: 5’- GACGUCCAGUCACTUGTCGCA-3’.
Enzyme-Linked Immunosorbent Assay (ELISA)
EGFR ELISA kit (RAB0160), IL-10 ELISA kit (RAB0244), TGF-β ELISA kit (RAB0460), IL-1β ELISA kit (RAB0273), and CCL18 ELISA kit (RAB0051) were from Sigma-Aldrich (Germany). The protein level in the cell culture supernatant was detected according to the manufacturer’s protocol.
RNA Isolation and Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
Total cellular RNA was extracted by TRIzol (Beyotime, China). qRT-PCR assay was performed with QuantiTect SYBR Green PCR Kit (Qiagen, Germany) on Stepone Real-Time PCR System (Applied Biosystems, USA). The primer sequences are presented in Table 2, which were synthesized from Genewiz (Genewiz, Tianjin, China). The data were analyzed by the 2−ΔΔCt method (Biorad CFX manager software 3.1).
 |
Table 2 Primer Sequences |
Cell Viability Detection
Cell viability was measured by a CCK8 kit (Abcam, Britain). Briefly, 3000 cells per well were seeded in a 96-well plate (Corning, USA). After incubation, the culture solution was discarded, and 10 μL CCK-8 solution was added to each well. Then cells were protected from light for 1 h at 37°C during incubation, and cellular proliferation was estimated by a microplate reader (Bio-Tek, USA).
Animal Experiments
Animal experiments were approved by the Institutional Animal Care and Use Committee of Beijing Viewsolid Biotechnology Co., LTD and followed the Laboratory Animal—Guideline for Ethical Review of Animal Welfare (GB/T 35892-2018). The xenograft mouse model of esophageal cancer was developed to observe tumor growth in vivo. NOD-SCID mice (female, six-week-old) were used in all animal experiments. For detecting the effect of the co-culture medium of macrophages and exosomes on tumor growth, the mice were randomly divided into four groups (n=4). All mice were injected subcutaneously with 2×106 wild OE33 cells in the flank region. From the following day, the control group was injected with 100 μL RPMI 1640 medium, and the other three groups were injected with 100 μL different co-culture medium daily. To verify the effects of CCL18 and PITPNM3 on tumor growth, the mice were randomly divided into three groups (n=4). Firstly, the group 1 and group 2 were injected with OE33 cells stably expressing scrambled shRNA (2 × 106 cells in 200 μL PBS per group), and the group 3 was injected with OE33 cells stably expressing PITPNM3 shRNA (2 × 106 cells in 200 μL PBS). From the following day, the group 1 was injected intravenously with 100 μL PBS, and the group 2 and group 3 were injected with CCL18 recombinant protein solution (50 μg/kg) daily. Tumor volumes were measured weekly. After 4 weeks, the tumors were excised. Tumor volume was calculated using this formula: 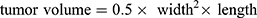 .
.
Immunohistochemical Staining
Tumor tissues were collected and fixed in 10% phosphate-buffered formalin, paraffin-embedded, and sectioned. After a series of treatments, sections were blocked with 5% BSA, incubated with anti-Ki67 antibody or anti-PITPNM3 antibody for 3h, and then incubated with the HRP-conjugated secondary antibody for 3h at room temperature. Finally, counterstaining was performed with hematoxylin. Images were taken using a light microscope.
Statistical Analysis
Data are expressed as the mean±standard deviation (SD) from at least three independent experiments. Statistical analysis for the data was performed with GraphPad Prism 9.3.0. A normal distribution test was first conducted, and the Levene method was used to test the uniformity of square difference. The one-way analysis of variance and the Student’s unpaired t-test were used to compare the difference between multiple groups and two groups, respectively. The F-value was calculated. A p<0.05 suggested a significant difference.
Results
ABS Improve the Exosome-Delivered EGFR of Normal Esophageal Epithelial Cell, BE Cell, and EACcell
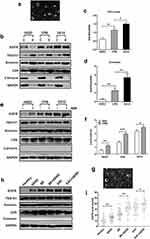
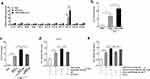
Firstly, we detected the EGFR level in the whole cell lysates and exosomes of normal esophageal epithelial cell line HEEC, BE cell line CPB, and EAC cell line OE33, respectively. Figure 1a shows a cell line-derived exosome representative electron microscopy picture. For western-blot experiments, TSG101, Annexin, and CD9 were used to be positive markers of the exosomes, and calnexin was used to be a negative marker. As shown in Figure 1b–d, the EGFR level increased gradually with an increasing degree of cell malignancy in both cell lysate and exosome. Afterward, we treated HECC, CPB, and OE33 cells with an ABS cocktail (100 μM, pH 4) for 36h. The result indicated that ABS significantly improved EGFR expression in the exosomes from different cell lines (Figure 1e and f).
Consequently, we extracted the exosomes from the serum collected from healthy adults and patients suffering from GERD, BE with or without GERD, and EAC with or without GERD, respectively. Figure 1g shows a representative electron microscopy picture of the serum-derived exosome. As shown in Figure 1h and i, the expression of exosome-delivered EGFR increased as increasing disease severity. We found that in BE or EAC patients, the expression of EGFR in patients with GERD was higher than that in patients without GERD. The results are consistent with our results from cell lines, which indicates that gastroesophageal reflux fluid (acidic bile salts are the main components) maybe a vital factor for increasing the exosome-delivered EGFR and further aggravating disease progression.
Exosomes from ABS-Treated Cells Promote the M2 Polarization of Macrophages by Increasing Exosome-Delivered EGFR Levels
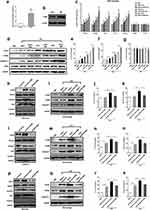
Next, we detected the effect of exosomes from the normal esophageal epithelial cell, Barrett’s esophageal cell, or esophageal adenocarcinoma cell on the proliferation of their corresponding cells. Nevertheless, no significant results were observed (data not shown). Then, we wanted to determine whether the exosomes could regulate the function of other cells in TME. It is well known that TAMs, as the main cellular components of the TME, play essential roles in tumor progression.15 Therefore, we investigated whether exosomes could regulate macrophage polarity. At first, resting macrophages (M0) were achieved by treating human THP-1 monocytes with 100 ng/mL PMA, which was demonstrated by the increased macrophage marker CD68 (Figure 2a and b). Then, we used the exosomes from different cell lines to treat M0 macrophages. After that, the markers of M1 macrophage including iNOS and interleukin-1beta (IL-1β), and the markers of M2 macrophage including CD206, CD163, arginase-1 (Arg1), interleukin-10 (IL-10) and transforming growth factor (TGF-β) were measured. The results showed that the expression of M2 macrophage markers in both mRNA (Figure 2c) and protein (Figure 2d–g) levels were improved with increasing degree of cell malignancy and was further promoted by the exosomes from the cells treated with ABS; meanwhile, the expression level of M1 macrophage markers did not change. In addition, the exosome from HEEC did not affect the M2 polarization of macrophages.
From the above results, the M2 polarization trend was consistent with the level of EGFR in the exosome, so we speculated that EGFR might be involved in regulating M2 polarization, and the enhancement effect of ABS might depend on exosome-derived EGFR level. Therefore, we decreased EGFR expression of HEEC, CPB, and OE33 cell lines by siRNA transfection to demonstrate the effect of EGFR expression induced by ABS on macrophage M2 polarization. As shown in Figure 2h, the results indicated that the expression of EGFR induced by ABS in exosomes from HEEC was decreased by EGFR knockdown. In Figure 2i–k, the knockdown of EGFR remarkably reduced macrophage M2 polarization induced by exosomes from ABS-treated HEEC cells. Similarly, EGFR knockdown also decreased macrophage M2 polarization induced by the exosome from ABS-treated CPB (Figure 2i–o) and OE33 cells (Figure 2p–s), respectively. These results indicate that ABS improve exosome-delivered EGFR in all cell lines, further promoting the M2 polarization of macrophage.
Exosome-Delivered EGFR Regulates Macrophage M2 Polarization by PI3K/AKT Pathway
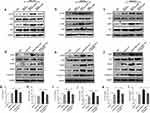
Furthermore, the detailed mechanism of exosome-delivered EGFR regulating macrophage M2 polarization was investigated. It has been known that EGFR/PI3K/AKT signaling pathway is involved in regulating various cellular functions, including cell proliferation,25 M2 polarization,26 and other cell processes. As shown in Figure 3a–c, compared with the exosomes from non-ABS-treated cells, we found that the exosomes from ABS-treated HEEC, CPB, and OE33further promoted the phosphorylation of PI3K and AKT in macrophages; however, EGFR knockdown in HEEC, CPB, and OE33 inhibited this process. These results demonstrate that ABS treatment promotes the activation of macrophage PI3K/AKT pathway by increasing exosome-delivered EGFR. To further verify whether this signaling pathway induces M2 polarization of macrophages, we used LY294002 (a PI3K/AKTpathway inhibitor) to treat macrophages. As shown in Figure 3d–l, we found thatLY294002 effectively suppressed the phosphorylation of AKT and the expression of M2 macrophage markers induced by the exosome from ABS-treated HEEC, CPB, and OE33, respectively. The results indicate that exosome-delivered EGFR improves M2 polarization of macrophage by PI3K/AKT pathway.
The Co-Culture Medium of Macrophage and Exosomes Promotes the Proliferation of the Normal Esophageal Epithelial Cell, BE Cell and EAC Cell
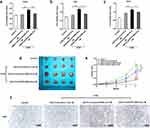
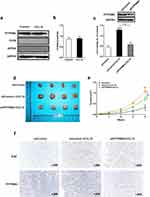
To further study whether macrophage M2 polarization can affect cell proliferation, we collected the co-culture medium of macrophages and exosomes from different cells (cells transfected with siControl, cells transfected with siControl and treated with ABS, and cells transfected with siEGFR and treated with ABS, respectively), and add them into HEEC, CPB and OE33 cells, respectively. A culture medium of macrophages cultured alone was used as a negative control. After incubation for the indicated time, the cell viability was measured. As presented in Figure 4a, HEEC cell proliferation was increased by the co-culture medium of macrophages and the exosome from HEEC pretreated by ABS, which was suppressed by exosome-delivered EGFR knockdown. The proliferation of CPB (Figure 4b) and OE33 cells (Figure 4c) showed similar results. Nevertheless, unlike the HEEC cell, the co-culture medium of macrophage and the exosome from CPB and OE33 cell also improved cell proliferation without ABS treatment. Subsequently, a murine tumor xenograft model was used to validate the co-culture medium’s effect on EAC tumor growth in vivo. The result indicated that the tumor growth was increased by the co-culture medium of macrophages and the exosome from OE33, which was further improved by ABS treatment. At the same time, the decreased exosome-delivered EGFR reduced tumor growth (Figure 4d and e). Ki67 staining of tumor sections also showed the similar results (Figure 4f).
M2 Macrophage Improving the Proliferation of EAC Cells by Releasing CCL18 Isexosome-Delivered EGFR Dependent
From the above results, the co-culture medium of macrophages with exosomes could strengthen the proliferation of normal esophageal epithelial cells, BE cells, orEAC cells, which suggested that macrophages may release some factors into the co-culture medium. Therefore, M2 macrophage-related cytokines were examined after macrophage incubation with exosome. The following experiments focused on the EAC cell lineOE33. As shown in Figure 5a, after the macrophage was incubated with exosome from OE33, the expressions of all cytokines were increased to different extents, including IL-10, TGF-β, vascular endothelial-derived growth factor (VEGF), Arg1, CCL1, CCL13, CCL16, CCL17, CCL18, CCL22, and CCL24. Among them, the mRNA expression of CCL18 was the most notably upregulated. Compared with exosomes from non-ABS treatment OE33 cells, the promotion effect of exosomes from ABS-treated OE33 cells on cytokines release was more potent. We further detected CCL18 concentration in the co-culture medium of macrophages with exosomes by ELISA. As shown in Figure 5b, compared with the PBS control group, the CCL18 level was elevated in the OE33-exosome treatment group, and the ABS treatment improved the effect. These results suggested that CCL18 may play a vital role in EAC cell proliferation. From the former results, we know that ABS could improve exosome-derived EGFR. To clarify the role of exosome-delivered EGFR in this process, we down-regulated EGFR expression in OE33 and found the CCL18 release reduced (Figure 5c). Consequently, the effect of CCL18 on EAC cell proliferation was investigated. After the co-culture medium of macrophages with exosomes was added into OE33, the viability of OE33 was examined. The result in Figure 5d indicated that the macrophage co-culture medium with exosome from OE33 improved the cell viability, and the exosome from ABS treatment OE33 showed a more substantial facilitation effect. However, CCL18 neutralizing antibody inhibited the effect of the co-culture medium. To further demonstrate the role of CCL18 in cell proliferation, we added CCL18 recombination protein into the co-culture medium, which rescued the decrease of OE33 cell proliferation induced by EGFR knockdown (Figure 5e).
CCL18 Promotes EAC Cell Proliferation and Tumor Growth by PITPNM3
It has been known that the receptor of CCL18 includes PITPNM3, CCR8, and GPR30, in which PITPNM3 is demonstrated as the most common one. To clarify the receptor of CCL18, we detected the expression of PITPNM3, CCR8, and GPR30 in EAC cell line OE33. As shown in Figure 6a and b, only PITPNM3 expression was determined in the OE33 cell line, and CCL18 binding could not change its expression level, which indicated that the OE33 cell constitutively expressed PITPNM3. We further investigated if PITPNM3 could mediate the function of CCL18 improving EAC cell growth. As presented in Figure 6c, PITPNM3 knockdown suppressed the increase of OE33 cell viability induced by CCL18. Finally, we constructed a murine xenograft tumor model by injection of OE33 cells transfected with PITPNM3 shRNA or shControl. As indicated in Figure 6d and e, the CCL18 recombination protein noticeably facilitated tumor growth. However, PITPNM3 knockdown inhibited the effect of CCL18. The result of Figure 6f further confirmed that CCL18 improved tumor growth via binding to PITPNM3.
Discussion
It is well known that GERD is an important risk factor for BE, and BE is a very significant link between GERD and EAC.27,28 Up to now, the specific mechanism behind this is still unclear. In this study, EGFR expression was found in cell lines (HEEC, CPB, and OE33), cell-derived exosomes, and serum-derived exosomes. The expression of EGFR increased gradually with an increasing degree of cell malignancy or increasing disease severity. EGFR plays a critical role in the homeostatic regulation of normal cells and the development of epithelial malignancies. It is a biomarker for cancer diagnosis and a therapeutic target in cancer therapy.29–32 The functions of EGFR in cancer cells have been well studied, but the roles of exosome-derived EGFR are less understood, especially in esophageal cancer. Here, our results indicated that higher EGFR expression was found in exosomes from cell lines with high EGFR expression. Figueroa et al found that cerebrospinal fluid-derived extracellular vesicles had significantly more EGFR expression in EGFR-positive glioblastomas,33 which is in line with our result. In this study, the EGFR content in a normal esophageal epithelial cell line or healthy person serum-derived exosome is low. Kharmate et al reported that exosome-delivered EGFR from healthy persons was relatively low,34 which is consistent with our results. In addition, it has been reported that the serum level of EGFR in non-small cell lung cancer patients is elevated and positively correlated with tumor progression.35 Interestingly, our results found a similar trend: the EGFR contents of serum-derived exosomes gradually increased from healthy persons to EAC patients. Moreover, we found that the ABS cocktail could improve the EGFR expression in all cell lines and cell-line-derived exosomes. GERD also further facilitated serum-derived exosomal EGFR expression of BE and EAC patients. Next, we wondered about the roles these exosomes can play. We first investigated the direct effect of exosomes on the proliferation of HEEC, CPB, and OE33 cell lines. However, no significant results were observed (data not shown). Then, we considered whether exosomes indirectly affect cellular functions through other manners, such as paracrine.
TAMs, as central components of TME, play crucial roles in regulating cancer initiation and development. There are many studies on tumor cell-derived exosomes promoting cancer progression by regulating the M1/M2 polarization of TAMs. For instance, highly accumulated circVCP in exosomes from the plasma of colorectal cancer (CRC) patients and CRC cells promotes the proliferation, migration, and invasion of cancer cells, stimulating macrophage M2 polarization.36 Besides, lung adenocarcinoma (LUAD) cell-derived exosomes induce M2 macrophage polarization by activating the JNK signaling pathway and further promoting LUAD progression.37 However, there are few studies on the roles of esophageal cancer-derived exosomes in TAMs polarization and cancer progression. Only one article showed that exosome-delivered miR-301a-3p from ESCC cells induced macrophage M2 polarization by activating PI3K/AKT signaling pathway to promote angiogenesis.38 The roles of EAC-derived exosomes in regulating ATMs functions during cancer progression are still unknown. In this study, the exosomes from BE and EAC cells induced the M2 polarization of macrophages, and ABS further improved the effect. However, the exosomes from the normal esophageal epithelial cell cannot induce macrophage M2 polarization, possibly due to EGFR’s low expression in exosomes. ABS treatment increased the exosomal EGFR levels of normal esophageal epithelial cell and further improved macrophage M2 polarization. A previous study reported that EGFR was abundantly expressed in cancer cell-delivered exosomes, which promoted the survival of monocytes (the precursors of ATMs) by activating the MAPK pathway,14 suggesting that the expression level of exosomal EGFR may be tightly associated with the function of ATMs. PI3K/AKT is the main downstream signaling pathway of EGFR. The exosomal EGFR in NSCLC activates PI3K/AKT signaling pathway and induced osimertinib resistance.39 ESCC cell-derived exosomes induce macrophage M2 polarization by activating PI3K/AKT signaling pathway.38 Here, OE33-derived exosomes activated PI3K/AKT pathway to improve the M2 polarization of macrophages. ABS cocktail further improved the effect by increasing exosomal EGFR expression.
Subsequently, we found that the co-culture medium of macrophage and exosomes promoted the proliferation of HEEC, CPB, and OE33 cell lines. ABS cocktail treatment further enhanced the proliferation, which was prevented by EGFR knockdown. The tumor growth showed a similar result in a murine tumor xenograft model. The results suggested that macrophages may secret some factors into the cell culture medium after co-culture with exosomes, and this secretion might be related with exosomal EGFR expression induced by the ABS cocktail. It has been known that M2 macrophages can release various cytokines under different conditions to regulate TME and play essential roles in cancer initiation and progression.40 For instance, CCL22 and IL-1β released from M2 macrophages can improve ESCC cell migration and invasion.41 In TAM samples isolated from breast carcinomas patients, CCL18 is the most abundantly expressed cytokines and correlated with larger tumor size, higher grade, and lymph node or distal metastasis.42 Moreover, CCL18 is expressed at high levels in ESCC tissues, which is related to worse survival of ESCC patients, and CCL18 can also improve the invasive ability of ESCC cells in a dose-dependent manner.43 Therefore, after the macrophages were treated by OE33 cell line-derived exosome, we detected the expressions of several most common M2 macrophage related-cytokines. We were surprised to find that the mRNA expression of CCL18 was upregulated with the most significant fold change and remarkably higher than other cytokines. Next, more results indicated that the increase of CCL18 was dependent on exosome-delivered EGFR induced by the ABS cocktail, and CCL18 was also demonstrated to be the major component in promoting cell proliferation. PITPNM3 is the most common receptor of CCL18. CCL18 promotes the cell migration, invasion, and EMT of PITPNM3-positive hepatocellular carcinoma cells.44 In addition, PITPNM3 silence reverses the effect of CCL18 on the proliferation, migration, and invasion of intrahepatic cholangiocarcinoma.45 However, in esophageal cancer, the roles of CCL18 and PITPNM3 in cancer progression are unknown. Here, the PITPNM3 is determined to be the receptor of CCL18 in the OE33 cell line. It is constitutively expressed in the OE33 cell line and mediates the cell proliferation and xenografted tumor growth induced by CCL18. CCR8 and GPR30, the other two receptors of CCL18, are not detected in OE33 cells. In the future, the downstream signaling pathway of CCL18 regulating EAC proliferation via PITPNM3 should be investigated. The effect of ABS on EAC invasion and metastasis also needs to study. Moreover, this study is mainly focused on the effect of the cytokine released by M2 macrophage on the EAC proliferation and xenografted tumor growth. But, the details of the interaction between M2 macrophage and normal human esophageal epithelial cell or BE cell are still unknown. This question merits further investigation.
Conclusions
In summary, our study indicates that ABS improve exosome-delivered EGFR, which induces macrophage M2 polarization by activating the PI3K/AKT signaling pathway. CCL18 released from the M2 macrophage binds to PITPNM3 and promotes EAC cell proliferation. These findings provide new evidence to understand the roles of ABS in EAC progression.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Informed Consent
The human ethics and research ethics committees of Tianjin Medical University Cancer Institute and Hospital approved the study (approval no. bc2020176).
Funding
This work was supported by the National Key Clinical Specialist Construction Programs of China (No. 2013-544), National Natural Science Foundation of China (82072059 and 82172090) and Tianjin Key Medical Discipline (Specialty) Construction Project (No. TJYXZDXK-010A).
Disclosure
The authors declare no competing interests.
References
1. Uhlenhopp DJ, Then EO, Sunkara T, Gaduputi V. Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol. 2020;13(6):1010–1021.
2. Mejza M, Malecka-Wojciesko E. Diagnosis and management of barrett’s esophagus. J Clin Med. 2023;12(6):2141. doi:10.3390/jcm12062141
3. Chen L, Lu H, Peng D, et al. Activation of NOTCH signaling via DLL1 is mediated by APE1-redox-dependent NF-kappaB activation in oesophageal adenocarcinoma. Gut. 2023;72(3):421–432. doi:10.1136/gutjnl-2022-327076
4. Storz L, Walther P, Chemnitzer O, et al. Nrf2/Keap1-pathway activation and reduced susceptibility to chemotherapy treatment by acidification in esophageal adenocarcinoma cells. Cancers. 2021;13(11):2806. doi:10.3390/cancers13112806
5. Liu J, Liu L, Su Y, et al. IL-33 participates in the development of esophageal adenocarcinoma. Pathol Oncol Res. 2022;28:1610474. doi:10.3389/pore.2022.1610474
6. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478). doi:10.1126/science.aau6977
7. Seneff S, Nigh G, Kyriakopoulos AM, McCullough PA. Innate immune suppression by SARS-CoV-2 mRNA vaccinations: the role of G-quadruplexes, exosomes, and MicroRNAs. Food Chem Toxicol. 2022;164:113008. doi:10.1016/j.fct.2022.113008
8. Huang TY, Wang CY, Chen KY, Huang LT. Urinary exosomal thyroglobulin in thyroid cancer patients with post-ablative therapy: a new biomarker in thyroid cancer. Front Endocrinol. 2020;11:382. doi:10.3389/fendo.2020.00382
9. Bano R, Soleja N, Mohsin M. Genetically encoded FRET-based nanosensor for real-time monitoring of A549 exosomes: early diagnosis of cancer. Anal Chem. 2023;95(13):5738–5746. doi:10.1021/acs.analchem.2c05774
10. Rajaram P, Chandra P, Ticku S, Pallavi BK, Rudresh KB, Mansabdar P. Epidermal growth factor receptor: role in human cancer. Indian J Dent Res. 2017;28(6):687–694. doi:10.4103/ijdr.IJDR_534_16
11. Zhang H, Deng T, Liu R, et al. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat Commun. 2017;8(1):15016. doi:10.1038/ncomms15016
12. Zhou Z, Wu X, Zhan R, et al. Exosomal epidermal growth factor receptor is involved in HPV-16 E7-induced epithelial-mesenchymal transition of non-small cell lung cancer cells: a driver of signaling in vivo? Cancer Biol Ther. 2022;23(1):1–13. doi:10.1080/15384047.2022.2133332
13. Huang S-H, Li Y, Zhang J, Rong J, SJCi Y. Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Invest. 2013;31(5):330–335.
14. Song X, Ding Y, Liu G, et al. Cancer cell-derived exosomes induce mitogen-activated protein kinase-dependent monocyte survival by transport of functional receptor tyrosine kinases. J Biol Chem. 2016;291(16):8453–8464. doi:10.1074/jbc.M116.716316
15. Jayasingam SD, Citartan M, Thang TH, Mat Zin AA, Ang KC, Ch’ng ES. Evaluating the polarization of tumor-associated macrophages into M1 and M2 phenotypes in human cancer tissue: technicalities and challenges in routine clinical practice. Front Oncol. 2019;9:1512. doi:10.3389/fonc.2019.01512
16. Pittet MJ, Michielin O, Migliorini D. Clinical relevance of tumour-associated macrophages. Nat Rev Clin Oncol. 2022;19(6):402–421. doi:10.1038/s41571-022-00620-6
17. Ruytinx P, Proost P, Van Damme J, Struyf S. Chemokine-induced macrophage polarization in inflammatory conditions. Front Immunol. 2018;9:1930. doi:10.3389/fimmu.2018.01930
18. Wang X, Luo G, Zhang K, et al. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kγ to promote pancreatic cancer metastasis tumor-promoting effects of hypoxic exosomal miR-301a. Cancer Res. 2018;78(16):4586–4598. doi:10.1158/0008-5472.CAN-17-3841
19. Cheng Y, Zhong X, Nie X, et al. Glycyrrhetinic acid suppresses breast cancer metastasis by inhibiting M2-like macrophage polarization via activating JNK1/2 signaling. Phytomedicine. 2023;114:154757. doi:10.1016/j.phymed.2023.154757
20. Weinhauser I, Pereira-Martins DA, Almeida LY, et al. M2 macrophages drive leukemic transformation by imposing resistance to phagocytosis and improving mitochondrial metabolism. Sci Adv. 2023;9(15):eadf8522. doi:10.1126/sciadv.adf8522
21. Li J, Xie Y, Wang X, et al. Prognostic impact of tumor-associated macrophage infiltration in esophageal cancer: a meta-analysis. Future Oncol. 2019;15(19):2303–2317. doi:10.2217/fon-2018-0669
22. Miyashita T, Tajima H, Shah FA, et al. Impact of inflammation-metaplasia-adenocarcinoma sequence and inflammatory microenvironment in esophageal carcinogenesis using surgical rat models. Ann Surg Oncol. 2014;21(6):2012–2019. doi:10.1245/s10434-014-3537-5
23. Cao W, Peters JH, Nieman D, Sharma M, Watson T, Yu J. Macrophage subtype predicts lymph node metastasis in oesophageal adenocarcinoma and promotes cancer cell invasion in vitro. Br J Cancer. 2015;113(5):738–746. doi:10.1038/bjc.2015.292
24. Bhat AA, Lu H, Soutto M, et al. Exposure of Barrett’s and esophageal adenocarcinoma cells to bile acids activates EGFR–STAT3 signaling axis via induction of APE1. Oncogene. 2018;37(46):6011–6024. doi:10.1038/s41388-018-0388-8
25. Shi C, Sun L, Fang R, Zheng S, Yu M, Li Q. Saikosaponin-A exhibits antipancreatic cancer activity by targeting the EGFR/PI3K/Akt pathway. Curr Pharm Biotechnol. 2023;24(4):579–588. doi:10.2174/1389201023666220610113514
26. Lian G, Chen S, Ouyang M, Li F, Chen L, Yang J. Colon cancer cell secretes EGF to promote M2 polarization of TAM through EGFR/PI3K/AKT/mTOR pathway. Technol Cancer Res Treat. 2019;18:1533033819849068. doi:10.1177/1533033819849068
27. Manner H. Update Barrett-Ösophagus [Current status in Barrett’s esophagus]. Dtsch Med Wochenschr. 2023;148(3):93–102. German. doi:10.1055/a-1832-3984
28. Lagergren J. Oesophageal cancer and gastro-oesophageal reflux: what is the relationship? Gut. 2004;53(8):1064–1065. doi:10.1136/gut.2003.038471
29. Jenkins R, Walker J, Roy UB. Plain language summary and patient perspective of the European society for medical oncology expert consensus statements on treating EGFR-positive non-small-cell lung cancer. Future Oncol. 2023;2023:1.
30. Peng W, Yao C, Pan Q, et al. Novel considerations on EGFR-based therapy as a contributor to cancer cell death in NSCLC. Front Oncol. 2023;13:1120278. doi:10.3389/fonc.2023.1120278
31. Karan C, Tan E, Sarfraz H, et al. Human epidermal growth factor receptor 2-targeting approaches for colorectal cancer: clinical implications of novel treatments and future therapeutic avenues. JCO Oncol Pract. 2022;18(8):545–554. doi:10.1200/OP.21.00904
32. Wu Y, Zeng X, Gan Q. A compact surface plasmon resonance biosensor for sensitive detection of exosomal proteins for cancer diagnosis. Methods Mol Biol. 2022;2393:3–14.
33. Figueroa JM, Skog J, Akers J, et al. Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro Oncol. 2017;19(11):1494–1502. doi:10.1093/neuonc/nox085
34. Kharmate G, Hosseini-Beheshti E, Caradec J, Chin MY, Tomlinson Guns ES, Nie D. Epidermal growth factor receptor in prostate cancer derived exosomes. PLoS One. 2016;11(5):e0154967. doi:10.1371/journal.pone.0154967
35. Cao X, Zhong W, Guo S, Zhang Z, Xie C. Low expression of miR-27b in serum exosomes of non-small cell lung cancer facilitates its progression by affecting EGFR. Open Med. 2022;17(1):816–825. doi:10.1515/med-2022-0472
36. Tang Y, Hu S, Li T, Qiu X. Tumor cells-derived exosomalcircVCP promoted the progression of colorectal cancer by regulating macrophage M1/M2 polarization. Gene. 2023;870:147413. doi:10.1016/j.gene.2023.147413
37. Xu L, Wang L, Yang R, Li T, Zhu X. Lung adenocarcinoma cell-derived exosomes promote M2 macrophage polarization through transmission of miR-3153 to activate the JNK signaling pathway. Hum Mol Genet. 2023;32(13):2162–2176. doi:10.1093/hmg/ddad052
38. Shou Y, Wang X, Chen C, et al. Exosomal miR-301a-3p from esophageal squamous cell carcinoma cells promotes angiogenesis by inducing M2 polarization of macrophages via the PTEN/PI3K/AKT signaling pathway. Cancer Cell Int. 2022;22(1):153. doi:10.1186/s12935-022-02570-6
39. Wu S, Luo M, To KKW, et al. Intercellular transfer of exosomal wild type EGFR triggers osimertinib resistance in non-small cell lung cancer. Mol Cancer. 2021;20(1):17. doi:10.1186/s12943-021-01307-9
40. Mantovani A, Allavena P, Marchesi F, Garlanda C. Macrophages as tools and targets in cancer therapy. Nat Rev Drug Discov. 2022;21(11):799–820. doi:10.1038/s41573-022-00520-5
41. Zhou J, Zheng S, Liu T, et al. IL-1beta from M2 macrophages promotes migration and invasion of ESCC cells enhancing epithelial-mesenchymal transition and activating NF-kappaB signaling pathway. J Cell Biochem. 2018;119(8):7040–7052. doi:10.1002/jcb.26918
42. Chen J, Yao Y, Gong C, et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell. 2011;19(4):541–555. doi:10.1016/j.ccr.2011.02.006
43. Wang W, Wu D, He X, et al. CCL18-induced HOTAIR upregulation promotes malignant progression in esophageal squamous cell carcinoma through the miR-130a-5p-ZEB1 axis. Cancer Lett. 2019;460:18–28. doi:10.1016/j.canlet.2019.06.009
44. Lin Z, Li W, Zhang H, et al. CCL18/PITPNM3 enhances migration, invasion, and EMT through the NF-kappaB signaling pathway in hepatocellular carcinoma. Tumour Biol. 2016;37(3):3461–3468. doi:10.1007/s13277-015-4172-x
45. Wang C, Liang H, Li Y, Tang Z, Zhang Y. Chemokine (C-C motif) ligand 18/membrane-associated 3/forkhead box O1 axis promotes the proliferation, migration, and invasion of intrahepatic cholangiocarcinoma. Bioengineered. 2022;13(5):12738–12748. doi:10.1080/21655979.2022.2069383
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.