Back to Journals » Neuropsychiatric Disease and Treatment » Volume 16
Exosomal Long Non-Coding RNA Expression from Serum of Patients with Acute Minor Stroke
Authors Xu X, Zhuang C, Chen L
Received 9 September 2019
Accepted for publication 23 December 2019
Published 13 January 2020 Volume 2020:16 Pages 153—160
DOI https://doi.org/10.2147/NDT.S230332
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yuping Ning
Xiaonan Xu, 1,* Chengle Zhuang, 2,* Liming Chen 1
1Department of Neurology, Shenzhen University General Hospital, Shenzhen 518000, Guangdong Province, People’s Republic of China; 2Department of Urology, Peking University Shenzhen Hospital, Shenzhen 518000, Guangdong Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Liming Chen Email [email protected]
Background: Acute minor stroke (AMS) is one kind of hypoxic ischemic necrosis with no more than 4 National Institutes of Health Stroke Scale (NIHSS) score. However, the early diagnosis of AMS is tough for lack of effective molecular markers. Recently, many long non-coding RNAs (lncRNAs) associated with AMS have been gradually revealed. Here, we aim to find the potential biomarkers of lncRNAs in exosomes isolated from blood serum of patients with AMS for early detection.
Methods: RNA-seq technique, KEGG pathway analysis and GO enrichment analysis were used in this study. Besides, reverse transcription real-time quantitative polymerase chain reaction (RT-qPCR) was used to validate expression levels of four of eleven differentially expressed lncRNAs (lnc-CRKL-2, lnc-NTRK3-4, RPS6KA2-AS1 and lnc-CALM1-7) involved in the neurotrophin signaling pathway.
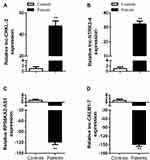
Results: The expression levels of lnc-CRKL-2 (mean value 48, standard deviation 4.583, P = 0.003) and lnc-NTRK3-4 (mean value 32.3, standard deviation 2.08, P = 0.001) were increased significantly in AMS patients, while the expression levels of RPS6KA2-AS1 (mean value − 118.7, standard deviation 7.09, P = 0.001) and lnc-CALM1-7 (mean value − 148.7, standard deviation 6.10, P = 0.001) were decreased dramatically.
Conclusion: In conclusion, these four new revealed lncRNAs may be used as novel joint biomarkers for the early detection of AMS.
Keywords: exosomes, stroke, LncRNAs, biomarker
Introduction
Acute stroke is a serious complication of cerebral vascular disease with a low occurrence of disability, which has been traditionally defined as NIHSS score with no more than 4.1,2 According to the increasing number of stroke patients, this disease becomes a severe public health concern owing to the first leading cause of mortality and disability.3,4 Furthermore, previous evidence have suggested that a number of severe major stroke patients suffer a stroke attack followed by initial minor stroke within the first few hours or days.5 The images of CT scan for acute minor stroke (AMS) are mostly normal and MR examination is expensive and time-consuming. At present, the diagnosis of minor stroke within 24-hr duration mainly relies on symptoms and signs. Hence, early detection, diagnosis and intervention are of great value for AMS patients.
Recently, lots of studies have shown that exosomes may function as intercellular communicators in the human body.6,7 Exosomes are considered as “cell dust” in the past while new studies show that many stable circulating potential biomarkers exist in exosomes.8 These potential biomarkers include DNAs,9 mRNAs/non-coding RNAs10 and proteins.11 The potential clinical application of exosomes is tremendously extensive.12,13 Exosomes isolated from disease-related biological samples were widely studied in order to improve diagnostic accuracy, especially in cancer, neuro-degenerative diseases and acute organ injury.14 In neuro-degenerative diseases, exosomes have been proven to possess potential diagnostic value in Alzheimer’s disease and Parkinson’s disease.15,16 However, no study demonstrates the potential value of exosomes in stroke disease.
Exosomes contain nucleic acid which major forms are RNAs.17 Studies have demonstrated that RNAs could be transferred to recipient cells and maintain their biological functions.18 Long non-coding RNAs (lncRNAs) are one kind of RNAs that are proven to exist in exosomes.14 LncRNAs mediate several vital pathways in the progression of stroke.19,20 For example, lncRNA SNHG14 can promote microglia activation through regulating miR-145-5P/PLA2G4A in stroke.21 In addition, lncRNA MALAT1 regulates cerebrovascular pathologies and neuronal cell death in stroke.22,23 Thus, lncRNAs may be potential molecules for the detection of stroke.
In this study, we used the RNA-seq technique to illustrate the lncRNA expression profiles in exosomes isolated from the blood serum of patients with AMS and healthy controls (HCs). And we explored the roles of lncRNAs in AMS using bioinformatics method and we validated the expression of four of the eleven lncRNAs (lnc-CRKL-2, lnc-NTRK3-4, RPS6KA2-AS1, lnc-CALM1-7) involved in neurotrophin signaling pathway through RT-qPCR. Thus, the new four lncRNAs might be utilized as potential circulating biomarkers of AMS for clinical early diagnosis.
Methods
Sample Collection
One hundred patients with acute minor stroke (AMS) and one hundred healthy controls were recruited from May 2014 to November 2018 of Peking University Shenzhen Hospital. All patients with AMS symptoms within 24 hrs have no any treatment before obtaining blood samples. AMS was diagnosed by an acute neurological impairment persistently for NIHSS≤4, also by a lesion on diffusion-weighted magnetic resonance subsequently. AMS patients and HCs were excluded from a medical history of cardiovascular/cerebrovascular diseases, chronic kidney/liver diseases, active malignant disease and inflammatory diseases.
Vacuum blood collection tubes were used to collect about 15 mL blood from AMS patients or HCs. The serum was isolated using centrifuge and then used for exosomes extraction. Informed consent was obtained from all patients or HCs and this study protocol was approved by the ethics committee of Shenzhen University General Hospital and Peking University Shenzhen Hospital. Written informed consents from the patients and healthy controls were signed, and this study was conducted in accordance with the Declaration of Helsinki.
Exosomes Extraction
ExoQuick-TCTM Exosome Precipitation Solution kit (System Bioscience, Mountain View, CA) was used for exosome extraction following the manual’s instructions: Collect 250μL serum from patients or healthy controls and centrifuge at 3000 × g for 15 min to remove cells and cell debris. Then, transfer the supernatant to a sterile vessel and add 63μL ExoQuick Exosome Precipitation Solution to it, and refrigerate for 12 hrs. The next day, centrifuge ExoQuick/serum mixture at 1500 × g for 30 min. After centrifugation, the exosomes were deposited at the bottom of the vessel. Spin-down residual ExoQuick solution by centrifugation at 1500 × g for 5 min. Remove all traces of fluid by aspiration. Resuspend exosome pellet in 1/10 to 1/100 of original volume using sterile Phosphate Buffered Saline (PBS) and aliquot in cryogenic vials and store at −80°C for forthcoming experiments.
Western Blot Analysis
CD63 and Tsg101 were regarded as biomarkers in exosomes24 and detected by Western blot analysis. The protein concentration was measured by the bicinchoninic acid (BCA) quantification assay (Pierce Biotechnology, Rockford, IL, USA). Equal amounts of whole protein extract were electrophoresed on SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes using a semi-dry transfer cell (Bio-Rad Laboratories, Hercules, CA, USA). The membranes were incubated overnight with specific primary antibodies against CD63 (1:1000; Abcam, USA), Tsg101 (1:1000; Abcam, USA) and GAPDH (1:1000; Abcam, USA) after blocked with 5% nonfat dry milk. Horseradish peroxidase-conjugated secondary antibody (Amersham, Arlington Heights, IL) was used to incubate the blot for 1 hr at room temperature on a rocking platform. Finally, super signal chemiluminescence reagents (Thermo Fisher Scientific, Inc., Massachusetts, USA) was used to detect signal intensities.
RNA-Seq Analysis
ExoRNeasy Serum/Plasma Maxi Kit (Qiagen, Hilden, Germany) was used to extract total RNA from exosomes. Three healthy controls and three stroke patients were selected randomly for RNA-seq analysis. The VAHTS Toal RNA-seq (H/M/R) Libray Prep Kit for Illumina (Vazyme, Nanjing, China) was used to prepare strand-specific RNA-seq libraries. Other flow paths were similar to a previous study.25
Identification of Differentially Expressed lncRNAs
The reads were first mapped to the latest UCSC transcript set using Bowtie2 version 2.1.026, and for lncRNA expression analysis, we used the transcripts set form Lncipedia (http://www.lncipedia.org). TMM (trimmed mean of M-values) was used to normalize the gene expression. Differentially expressed lncRNAs were identified using the edgeR program (version 3.3.2).27 LncRNAs showing altered expression with P < 0.05 and more than 1.5-fold changes were considered differentially expressed.
Pathway Analysis of lncRNAs
The canonical pathway analysis was performed using Ingenuity Pathway Analysis (IPA). IPA computes scores for each network according to the fit of the set of supplied focus lncRNAs. These scores indicate the likelihood of focus genes that belong to a network versus those obtained by chance. A score >2 indicates a focus gene network was not generated by chance alone.
RNA Extraction and RT-qPCR
The TRIzol reagent (Invitrogen, Grand Island, NY, USA) was used to extract total RNA from exosomes isolated from one hundred AMS patients and one hundred HCs according to the manufacturer’s protocol. PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Dalian, China) was employed to synthesize the cDNA from total RNA. We validated the expression of the selected lncRNAs (lnc-CRKL-2, lnc-NTRK3-4, RPS6KA2-AS1, lnc-CALM1-7) associated with neurotrophin signaling pathway by RT-qPCR on the Roche LightCycler 480 Real-Time PCR System. GAPDH was used as the endogenous control to normalize the data. The comparative 2−ΔΔCT method was used to analyze the relative expression of these genes. All of the experiments were performed at least three times.
Data Analysis
Student’s test was used for the comparisons of continuous variables. χ2 test was performed to the comparisons of the clinical characteristics of AMS patients and categorical variables. Independent t-test was utilized to assess differential expression of lncRNAs between HCs and AMS patients. Fisher’s exact test with false discovery rate (FDR) option was used for calculating the significance of the canonical pathway. All statistical analyses were performed with SPSS 21.0 (IBM corporation, NY, USA). A two-sided value of P < 0.05 was considered statistically significant.
Results
Clinical Characteristics of AMS Patients
Totally, 200 participants (100 AMS patients and 100 HCs) were enrolled and their serum samples were collected. Table 1 shows that the baseline characteristics were no significant difference between AMS patients and HCs. Clinical characteristics of three AMS patients and three HCs which were chosen randomly for RNA-seq study were also shown no obviously difference (data not shown).
 |
Table 1 Clinical Characteristics of Acute Minor Stroke |
Validation of Exosomes Isolated from Patients or HCs
In order to certify the exosomes extracted from serum, we detected the expression of exosomal markers CD63 and Tsg101 that only exist in exosomes24 through Western blot analysis. As shown in Figure 1, CD63 and Tsg101 were all expressed in three patients or healthy controls. The result indicated that exosomes were isolated successfully and the extracts were suitable for the below experiments.
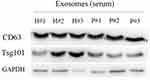 |
Figure 1 Western blot results demonstrate the expression of CD63 and Tsg101 in exosomes isolated from the serum of patients with acute minor stroke and healthy controls. |
Identification of Differentially Expressed lncRNAs
RNA-seq technique was used to detect differentially expressed lncRNAs in exosomes between the above selected 3 AMS patients and healthy controls. All 6733 lncRNAs detected between healthy controls and AMS patients were shown in Supplementary material 1. Heatmap of differential lncRNA expression was shown in Figure 2A. The results indicated that the expression levels of most lncRNAs in exosomes were down-regulated in AMS patients. Besides, the expression levels of 1096 lncRNAs changed with a P-value less than 0.05 (P < 0.05), including 307 upregulated lncRNAs and 789 downregulated lncRNAs (Figure 2B), and these differentially expressed lncRNAs were also listed in Supplementary material 1. Table 2 is the summary of the top 20 differentially expressed lncRNAs.
 |
Table 2 Top 20 Differentially Expressed lncRNAs in Exosomes Isolated from Serum of Patients with Acute Minor Stroke |
Analysis of Different lncRNAs That Involved in Signaling Pathways and Diseases
Most differentially expressed lncRNAs involved in focal adhesion, thyroid hormone signaling pathway, FOXO signaling pathway, autophagy, neurotrophin signaling pathway, and so on (Figure 2C–F). Neurotrophins have been shown to control many aspects of survival, development and function of neurons in nervous systems and may be novel targets for the design of stroke therapies.28,29 Thus, we will focus on neurotrophin signaling pathway in this study. In particular, eleven differentially expressed lncRNAs involved in neurotrophin signaling pathway (Figure 2C) are listed in Table 3. We chose two most up-regulated lncRNAs (lnc-CRKL-2, lnc-NTRK3-4) and two most down-regulated lncRNAs (RPS6KA2-AS1, lnc-CALM1-7) as the potential biomarkers for the diagnosis of AMS in the next RT-qPCR validation.
 |
Table 3 Eleven Differentially Expressed lncRNAs Involved in Neurotrophin Signaling Pathway |
Validation for the Expression of lncRNAs
Data of RT-qPCR showed that the expression levels of lnc-CRKL-2 and lnc-NTRK3-4 (Figure 3A and B, both P < 0.001) were significantly higher in exosomes from serum of patients with AMS than those from healthy controls. The expression of lncRNA RPS6KA2-AS1 was markedly decreased in AMS patients compared with healthy controls (Figure 3C, P < 0.001). Besides, the expression of lnc-CALM1-7 was obviously downregulated and almost 100 times lower than that in healthy controls (Figure 3D, P < 0.001).
Discussion
In recent years, accumulative evidence have shown that exosomes carry different types of biomolecules and have great potential for clinical applications.30,31 Now, exosomes are used as circulating biomarkers for various diseases, including breast cancer,32 lung cancer,33 neuro-degenerative disease,16,34 and so on. As we know, stroke is a main serious public health problem3 and many researchers were focusing on seeking circulating biomarkers for early detection. The new finding may improve diagnosis accuracy and reduce social burden. However, no study has focused on the relationship between exosomes and stroke.
In this study, we first reported the expression of lncRNAs in exosomes isolated from the serum of patients with minor stroke. Three healthy controls and three patients with minor stroke were selected for RNA-seq analysis. We analyzed the lncRNA profile between exosomes from AMS patients and HCs. Results show that most differential expressed lncRNAs were downregulated in exosomes isolated from the patients' serum. KEGG pathway analysis and GO enrichment analysis further manifested that differential expressed lncRNAs involve in various signaling pathways, such as MAPK pathway, neurotrophin signaling pathway, and so on. In order to find potential biomarkers for stroke, we analysed lncRNAs significantly associated with stroke diseases. Studies show that neurotrophins control many aspects of survival, development and function of neurons in nervous systems and may be novel targets for the design of stroke therapies.28,29 In this study, eleven differentially expressed lncRNAs involved in neurotrophin signaling pathway and four important genes (two most up-regulated lncRNAs and two most down-regulated lncRNAs of these eleven lncRNAs) were chosen. The results of RT-qPCR validated that expression levels of lnc-CRKL-2 and lnc-NTRK3-4 were significantly increased. However, the expression levels of RPS6KA2-AS1 and lnc-CALM1-7 were obviously downregulated in exosomes in patients with acute minor patients compared with the HCs.
In this study, we hope that we can use the conventional specimen such as blood for the diagnosis of AMS. According to a recent study,35 researchers use Invitrogen Total Exosome Isolation Kits to isolate total exosomes from blood samples for less than 3 hrs. We will employ these Kits for the isolation of exosomes in our future work. Lots of differentially expressed lncRNAs were found in this study and we will try to investigate the possible mechanisms of these lncRNAs in AMS through bioinformatics analysis. Many studies have demonstrated that lncRNAs can bind to miRNAs and regulate gene expression indirectly. Recent studies reported that lncRNAs act as scaffolds for proteins to affect neural pathways. In the future, we will detect the expression of these lncRNAs in neural cells and investigate their possible mechanisms.
Our results revealed that IncRNAs may discriminate from AMS patients and healthy people. It is hard for both CT scan and MR examination to figure out lesion location of infarction in time in AMS patients within 24 hrs after onset. For these reasons, we considered IncRNAs might be kinds of ideal diagnostic biomarkers for acute minor stroke.
Conclusion
In summary, four lncRNAs (lnc-CRKL-2, lnc-NTRK3-4, RPS6KA2-AS1 and lnc-CALM1-7) were revealed through RNA-seq and verified by RT-qPCR in AMS patients. These four lncRNAs may provide a new method for the early detection of minor stroke patients within 24 hrs after onset.
Acknowledgments
We thank all the donors whose names were not included in the author list, but who participated in this program. This work was supported by Project of Shenzhen Health and Family Planning Commission (SZFZ2018066) and Department of Neurology of Shenzhen University General Hospital.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Gao HH, Gao LB, Wen JM. Correlations of MCP-1-2518A>G polymorphism and serum levels with cerebral infarction risk: a meta-analysis. DNA Cell Biol. 2014;33:522–530. doi:10.1089/dna.2013.2263
2. O’Donnell MJ, Xavier D, Liu L, et al. Risk factors for ischaemic and intracerebral haemorrhagic stroke in 22 countries (the INTERSTROKE study): a case-control study. Lancet. 2010;376:112–123. doi:10.1016/S0140-6736(10)60834-3
3. Sen S, Rabinstein AA, Elkind MS, Powers WJ. Recent developments regarding human immunodeficiency virus infection and stroke. Cerebrovasc Dis. 2012;33:209–218. doi:10.1159/000335300
4. Towfighi A, Saver JL. Stroke declines from third to fourth leading cause of death in the United States: historical perspective and challenges ahead. Stroke. 2011;42:2351–2355. doi:10.1161/STROKEAHA.111.621904
5. Zuin M, Rigatelli G, Faggian G, L’Erario R, Chinaglia M, Roncon L. Could advanced drug delivery systems be the future in cardiovascular revascularization medicine? Vascular. 2017;25:447–448. doi:10.1177/1708538116682912
6. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi:10.1038/nri855
7. Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi:10.1038/nri2567
8. Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262:9412–9420.
9. Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi:10.1038/ncomms1180
10. Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi:10.1038/ncb1596
11. Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–283. doi:10.1586/epr.09.17
12. van der Pol E, Boing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64:676–705. doi:10.1124/pr.112.005983
13. Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–948. doi:10.1016/j.bbagen.2012.03.017
14. Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New technologies for analysis of extracellular vesicles. Chem Rev. 2018;118:1917–1950. doi:10.1021/acs.chemrev.7b00534
15. Coleman BM, Hill AF. Extracellular vesicles–their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Semin Cell Dev Biol. 2015;40:89–96. doi:10.1016/j.semcdb.2015.02.007
16. Vella LJ, Hill AF, Cheng L. Focus on extracellular vesicles: exosomes and their role in protein trafficking and biomarker potential in Alzheimer’s and Parkinson’s disease. Int J Mol Sci. 2016;17:173. doi:10.3390/ijms17020173
17. Ravasi T, Suzuki H, Pang KC, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi:10.1101/gr.4200206
18. Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi:10.1038/ncb1800
19. Wu Z, Wu P, Zuo X, et al. LncRNA-N1LR enhances neuroprotection against ischemic stroke probably by inhibiting p53 phosphorylation. Mol Neurobiol. 2017;54:7670–7685. doi:10.1007/s12035-016-0246-z
20. Zhao M, Wang J, Xi X, Tan N, Zhang L. SNHG12 promotes angiogenesis following ischemic stroke via regulating miR-150/VEGF pathway. Neuroscience. 2018;390:231–240. doi:10.1016/j.neuroscience.2018.08.029
21. Qi X, Shao M, Sun H, Shen Y, Meng D, Huo W. Long non-coding RNA SNHG14 promotes microglia activation by regulating miR-145-5p/PLA2G4A in cerebral infarction. Neuroscience. 2017;348:98–106. doi:10.1016/j.neuroscience.2017.02.002
22. Guo D, Ma J, Yan L, et al. Down-regulation of lncRNA MALAT1 attenuates neuronal cell death through suppressing Beclin1-dependent autophagy by regulating mir-30a in cerebral ischemic stroke. Cell Physiol Biochem. 2017;43:182–194. doi:10.1159/000480337
23. Zhang X, Tang X, Liu K, Hamblin MH, Yin KJ. Long noncoding RNA Malat1 regulates cerebrovascular pathologies in ischemic stroke. J Neurosci. 2017;37:1797–1806. doi:10.1523/JNEUROSCI.3389-16.2017
24. Zhang Y, Kim MS, Jia B, et al. Hypothalamic stem cells control ageing speed partly through exosomal miRNAs. Nature. 2017;548:52–57. doi:10.1038/nature23282
25. Xu H, Gong Z, Shen Y, Fang Y, Zhong S. Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics. 2018;10:187–197. doi:10.2217/epi-2017-0109
26. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi:10.1038/nmeth.1923
27. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi:10.1093/bioinformatics/btp616
28. Vidaurre OG, Gascon S, Deogracias R, et al. Imbalance of neurotrophin receptor isoforms TrkB-FL/TrkB-T1 induces neuronal death in excitotoxicity. Cell Death Dis. 2012;3:e256. doi:10.1038/cddis.2011.143
29. Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi:10.1098/rstb.2006.1894
30. Thery C. Cancer: diagnosis by extracellular vesicles. Nature. 2015;523:161–162. doi:10.1038/nature14626
31. Zhang Y, Wang XF. A niche role for cancer exosomes in metastasis. Nat Cell Biol. 2015;17:709–711. doi:10.1038/ncb3181
32. Moon PG, Lee JE, Cho YE, et al. Identification of developmental endothelial locus-1 on circulating extracellular vesicles as a novel biomarker for early breast cancer detection. Clin Cancer Res. 2016;22:1757–1766. doi:10.1158/1078-0432.CCR-15-0654
33. Reclusa P, Taverna S, Pucci M, et al. Exosomes as diagnostic and predictive biomarkers in lung cancer. J Thorac Dis. 2017;9:S1373–S1382. doi:10.21037/jtd
34. Shi M, Liu C, Cook TJ, et al. Plasma exosomal alpha-synuclein is likely CNS-derived and increased in Parkinson’s disease. Acta Neuropathol. 2014;128:639–650. doi:10.1007/s00401-014-1314-y
35. Pan B, Qin J, Liu X, et al. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. 2019;10:1096. doi:10.3389/fgene.2019.01096
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.