Back to Journals » OncoTargets and Therapy » Volume 13
Exosomal-lncRNA DLEU1 Accelerates the Proliferation, Migration, and Invasion of Endometrial Carcinoma Cells by Regulating microRNA-E2F3
Authors Jia J, Guo S , Zhang D, Tian X, Xie X
Received 13 May 2020
Accepted for publication 24 July 2020
Published 25 August 2020 Volume 2020:13 Pages 8651—8663
DOI https://doi.org/10.2147/OTT.S262661
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr XuYu Yang
Jianjun Jia,1 Suiqun Guo,2 Dong Zhang,1 Xiaohui Tian,3 Xingmei Xie1
1Department of Obstetrics and Gynecology, The First Affiliated Hospital, Jinan University, Guangzhou City, Guangdong Province 510632, People’s Republic of China; 2Department of Obstetrics and Gynecology, The Third Affiliated Hospital, Southern Medical University, Guangzhou City, Guangdong Province, People’s Republic of China; 3Department of Obstetrics and Gynecology, The Seventh Affiliated Hospital, Sun Yat-Sen University, Shenzhen City, Guangdong Province, People’s Republic of China
Correspondence: Jianjun Jia
Department of Obstetrics and Gynecology, The First Affiliated Hospital, Jinan University, No. 613, West Huangpu Avenue, Tianhe District, Guangzhou City, Guangdong Province 510632, People’s Republic of China
Tel +86-20-38688357
Email [email protected]
Purpose: Long non-coding RNAs (lncRNAs) may act as oncogenes in several cancers, including endometrial carcinoma (EC). The purpose of the current study is to investigate the regulatory mechanism of exosomal-lncRNA deleted in lymphocytic leukemia1 (DLEU1) on EC.
Methods: The expression levels of lncRNA DLEU1, microRNA-381-3p and E2F Transcription Factor 3 (E2F3) in EC tissues or cells were detected using quantitative reverse transcription–polymerase chain reaction (qRT-PCR). We then analysed the proliferation, migration, and invasion of EC cells by performing the MTT assay, wound healing assay, and transwell invasion assay, respectively. Identification of exosomes was detected using Western blot assay. The uptake of exosomes was detected by a confocal microscope. The effects of exosomes on EC cells were investigated by construction of cell co-culture system. The interactions among DLEU1, miR-381-3p and E2F3 were confirmed using the dual-luciferase reporter (DLR) assay.
Results: LncRNA DLEU1 expression was highly up-regulated in EC tissues and cells. Knockdown of DLEU1 inhibited the proliferation, migration, and invasion of EC cells. Exosomes could be uptaken by the ambient EC cells. MiR-381-3p was a target of DLEU1 and was negatively modulated by DLEU1. Overexpression of miR-381-3p suppressed the proliferation, migration, and invasion of EC cells. Additionally, E2F3 was the target gene of miR-381-3p and was negatively modulated by miR-381-3p. Upregulation of miR-381-3p and down-regulation of E2F3 reversed the promoting effect of exosomal DLEU1 on EC cells.
Conclusion: Exosomal DLEU1 accelerates the development of EC by regulating the miR-381-3p/E2F3 axis, thus DLEU1 may act as a possible therapeutic target for treating EC.
Keywords: endometrial carcinoma, exosomes, lncRNA DLEU1, miR-381-3p, E2F3
Introduction
Endometrial cancer (EC) is common and fearful in females.1,2 The rate of EC-resulted death accounts for a large proportion in women-related cancers.3,4 At present, surgery and chemoradiotherapy are the main methods for treating EC patients at the early stage.5 Nevertheless, patients diagnosed at advanced stages often undergo a worse outcome.6,7 Therefore, understanding the mechanisms that underlie EC is critical for developing a new treatment strategy. Exosomes are a kind of extracellular membranous vesicle, containing DNAs and various forms of RNAs.8 They are often released into the micro-environment and act as vehicles in cell-to-cell communication.9,10 More importantly, exosomes are increasingly recognized as a liquid biopsy technique to aid in the diagnosis of malignancies.11,12 Thus, to investigate exosomes that contain some specific nucleic acids and proteins may be of great significance in clinical application of EC.
Long non-coding RNAs (lncRNAs) have been reported to be involved in tumorigenesis and progression of EC.13–15 Up-regulation of lncRNA CHL1-AS1 accelerates the proliferation and migration of EC cells,13 and overexpression of lncRNA CCAT1 promotes the growth of EC.15 The high expression of lncRNA 14,327.1 motivates the potential of EC cells in migration and invasion.14 Additionally, emerging evidences have been displayed that lncRNA deleted in lymphocytic leukemia1 (DLEU1) is also involved in the occurrence and progression of EC. Up-regulation of DLEU1 promotes the viability, migration, invasive ability of EC cells,17 and the progression of EC.18
Recently, researchers have found that microRNAs (miRNAs) serve as anti-tumor roles by regulating cell proliferation, apoptosis, metastasis and invasion.19,20 MiR-6076 and miR-361 have been reported to suppress the proliferation and migration of EC cells.13,16 MiR-152 can dampen the growth and metastasis of EC.19 Besides, miR-381 has been displayed to play an inhibitory role in EC.21 Up-regulation of miR-381 inhibits the proliferation and invasion of EC cells.21 Nevertheless, whether lncRNA DLEU1 regulates miR-381-3p on EC remains unclear.
It has been widely known that E2F Transcription Factor 3 (E2F3) is a member of the E2Fs family.22 As an oncogene, it plays an important role in cell proliferation and apoptosis.23–26 E2F3 can accelerate progression of ovarian cancer (OC),23 and gastric cancer (GC),25 and E2F3 knockdown inhibits the proliferation, migration and invasion of bladder cancer cells.24 Furthermore, E2F3 promotes the cell cycle and proliferation, eventually expediting the progression of EC.26 Thus, this study preliminarily explored the potentially modulatory role of DLEU1/miR-381-3p/E2F3 axis in EC.
In current study, the effect of exosomal DLEU1 on the progression of EC, and the regulatory mechanisms between DLEU1 and miR-381-3p/E2F3 axis were investigated. Findings of this study may provide a potential therapeutic target for EC.
Materials and Methods
Tissues Collection
Sixty-two EC patients (<50, n = 27; ≥50, n = 35) were selected in The First Affiliated Hospital, Jinan University from 2017 to 2019. The age range of the 62 patients was 35 to 66 years (mean age, 46.1±4.9 years). All EC patients were confirmed via histopathological examination. The inclusion criteria included first-time diagnosis and no prior history of radiotherapy, chemotherapy or other adjuvant therapies. The exclusion criteria included the presence of other types of malignant tumor and patients who had received EC treatment before admission. The tumor and adjacent normal tissues (within 2 cm of the tumors) of the patients were collected by surgery. Each patient of this study obtained the written informed consent. The protocols of this study were reviewed and approved by ethical committee in The First Affiliated Hospital, Jinan University (Approval ID: 2019-LSPK-009).
Cell Grouping and Transfection
The human EC cell lines (KLE, Ishikawa, RL95-2 and HEC-1A) were procured from Procell Life Science & Technology, Ltd (Wuhan, China). The human endometrial-stromal cell line (hESC) was procured from Otwo Biotech, Ltd (Shenzhen, China). The cells were grown in Roswell Park Memorial Institute (RPMI) 1640 medium containing 10% fetal bovine serum (FBS) at 37°C with 5% CO2. shRNA-negative control (sh-NC), shRNA-E2F3 (sh-E2F3) and shRNA-DLEU1-1/-2 (sh-DLEU1-1/-2) were procured from Sangon Biotech, Inc (Shanghai, China). Overexpression-DLEU1 (pcDNA-DLEU1), miR-381-3p mimics and their negative controls (pcDNA-NC, miR-NC) were all procured from Ribo Biotech, Ltd (Guangzhou, China). The cells were transfected with the aforementioned agents using a Lipofectamine RNAiMAX kit (Invitrogen, Carlsbad, CA, USA) for 48 h. At 48 h after transfection, the cells were harvested to perform the following experiments.
Isolation of Exosomes
After transfection, the EC cells were sequentially cultured in RPMI 1640 medium containing 10% FBS without exosomes at 37°C with 5% CO2. After 72 h of culture, the supernate and cell fragments were separated using centrifuges. The collecting supernatant was utilized to extract the exosomes using a GM™ Exosome Isolation Reagent kit (Geneseed Biology, Inc, Guangzhou, China) and the mixtures were incubated at 4°C for 30 min. Subsequently, the mixtures were centrifuged at 2000 g for 30 min. The obtained precipitates were the exosomes. The surface marker proteins of exosomes CD9 (1:1000; Abcam), CD63 (1:1000; Abcam) and CD81 (1:1000; Abcam) were used to identify the exosomes by Western blot.
Establishment of Cell Co-Culture System
The receptor Ishikawa cells (without transfection) were seeded into the basolateral chamber of Transwell. After 24 h, Ishikawa cells (donor cells) transfected with sh-E2F3, miR-381-3p mimics, pcDNA-DLEU1 and pcDNA-NC were seeded into the apical chamber of Transwell culture plate. All the aforementioned cells were seeded with 2 × 105 cells per well. Following co-culture for 24 h, the receptor cells and the exosomes were collected for extracting the total RNA.
Quantitative Reverse Transcription-PCR (qRT-PCR)
Total RNA was extracted using a TRIzol kit (Invitrogen, Inc.) and reversely transcribed into cDNA using the GoScript reverse transcription system (Promega, Madison, WI, USA). Then, the cDNA was subjected to qRT-PCR analysis. The thermocycling conditions were as follows: 94°C for 5 min, followed by 40 cycles at 94°C for 10 sec, 60°C for 40 sec and 72°C for 1 min. Glyceraldehyde 3-phosphate dehydrogenase (GADPH) was used as the internal reference. Gene expression was quantified using the 2−ΔΔCt method.
Cell Viability Assay
The viability of EC cells was detected by MTT assay. Cells were seeded into a 96-well plate with 2 × 105 cells per well. Subsequently, the cells were incubated for 24, 48, 72 and 96 h. Then, 20 µL MTT (GENECHEM, Inc, Shanghai, China) was added to each well at different time points. After that, the cells were incubated for another 2 h at 37°C. The viability (OD450) was analysed using a Multiskan Spectrum microplate reader (Thermo Fisher Scientific).
Wound Healing Assay
Cells (2 × 105) were seeded into 6-well plates and grown until 100% confluence in RPMI 1640 medium. Then, wounds on the cell monolayer were created using a pipette tip. After that, the cells were incubated for 24 h in a serum-free medium. At 0 h and 24 h, the measurement and imaging of wound closure were measured by AxioVision v4.7 software (Carl Zeiss Meditec, Dublin, CA, USA) under a microscope (magnification, × 200).
Transwell Invasion Assay
Transwell chamber (Becton, Dickinson and Company, FL, NJ, USA) with Matrigel coating was utilized to detect cell invasion. The upper chamber was seeded at a density of 2 × 105 cells per well in a serum-free medium. RPMI 1640 medium in the lower chamber containing 10% FBS was used as chemoattractant. After incubation for 3 h at 37°C with 5% CO2, the cells in the lower chamber were stained for 15 min with 0.5% crystal violet (Merck KGaA, Darmstadt, Germany) at room temperature. Light microscope (magnification, × 400) was used to count the stained cells in five randomly selected fields.
Tumor Xenografts in Nude Mice
The healthy female BALB/c nude mice (weighing 20 ± 2g) were procured from Cavens Lab, Ltd (Changzhou, China). At a controlled temperature of 20°C, the mice were maintained in sterile environment under a 12 h cycle (12 h for light and 12 h for dark). Subsequently, mice were divided into two groups: the sh-DLEU1-1 group and the sh-NC group (n = 5). Sh-DLEU1-1 or sh-NC was integrated into lentiviral vector and then transfected into Ishikawa cells. After that, the transfected Ishikawa cells (2 × 106 cells/100 ul, s.c.) were injected into the right flanks of the nude mice. Tumor volumes were measured every other week. Four weeks later, mice were anesthetized with pentobarbital sodium (50 mg/kg) and then sacrificed by cervical dislocation. The tumor xenograft was separated from mice and weighted. The study was performed in the Animal Experimental Center of The First Affiliated Hospital, Jinan University. All animal experiments were undertaken according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and with the approval of ethical committee in The First Affiliated Hospital, Jinan University (Approval ID: 2018-LSPK-009).
Fluorescence-Labeled Exosomes and Uptake of the Exosomes Assay
Based on the manufacturer’s instructions, the Exo-Green fluorescent staining kit PKH67 (Yanzai Biology, Ltd, Shanghai, China) was used to label the exosomes. In brief, the exosomes were re-suspended with PBS and mixed with Exo-Green, and then incubated for 10 min. Exo Quick-TC reagent was added to terminate the labeling reaction. After the mixture was centrifuged, the supernatant was discarded and exosomes were re-suspended with PBS for further use. Cells (1 × 105) were seeded into a 35-mm dish, after the cells adhered to the wall, the labeled exosomes with 100–150 μL were added, cultured for 24 h, and observed under a confocal microscope (magnification, × 400).
Western Blot Analysis
RIPA buffer containing protease inhibitors was used to extract proteins from cells, followed by detecting the protein concentrations using the BCA Protein Assay Kit (Abcam, Cambridge, MA, USA). Approximately 30 µg proteins were separated by 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred into polyvinylidene fluoride (PVDF) membrane. Membrane blocking was performed using 5% bovine serum albumin (BSA) at room temperature. Next, the membrane was incubated overnight at 4°C with primary antibodies against E2F3 (1:1000; Abcam). Then, tris-buffered saline Tween-20 (TBST) was used to wash the membranes for 3 times. Subsequently, at room temperature, the HRP-conjugated anti-mice IgG secondary antibody (1:3000; Santa Cruz, Waltham, MA, USA) was added to incubate for 1 h. β-actin served as the internal reference. The membrane was developed by Chemiluminescence reagents (Thermo Fisher Scientific) under a Gel-Pro analyzer (version 4.0, USA).
Dual-Luciferase Reporter Assay
The cloning 3ʹUTR sequences containing the binding site were inserted into pGL3 vector, thus constructing the wild-type vector. The mutant-type vector was constructed using a Phusion Site-Directed Mutagenesis Kit (Thermo Fisher Scientific). Wild-type/mutant-type vector and miR-381-3p mimics/miR-NC were co-transfected into the cells at 37°C for 48 h. Then, the cells were lysed. The collecting supernatant was utilized to measure the relative luciferase activity by a Dual-Luciferase Reporter Assay System (Promega).
Statistical Analysis
SPSS Statistics software (version 20.0, USA) was used to perform statistical analyses. Data were presented as the means ± standard deviation (SD). Student’s t-tests were used to assess the differences between two groups. One-way ANOVA followed by Tukey’s multiple comparisons test was used to evaluate the differences among multiple groups. Pearson’s correlation analysis was used to determine the correlation between the expression levels of miR-381-3p and DLEU1/E2F3 in EC tissues. P-value less than 0.05 indicated a statistically significant difference. All experiments were conducted in triplicate in at least three independent trials.
Results
LncRNA DLEU1 is Highly Expressed in EC Tissues and EC Cells
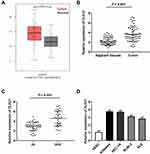
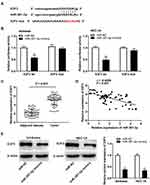
Based on TCGA database, DLEU1 expression in uterine corpus endometrial carcinoma (UCEC) and normal tissues was analysed. The results showed that DLEU1 expression was increased in UCEC tissues in contrast to that in normal tissues (Figure 1A, P < 0.05). The results of qRT-PCR displayed that DLEU1 expression was higher in tumor tissues in contrast to that in adjacent tissues (Figure 1B, P < 0.001). DLEU1 was highly expressed in stage III/IV of EC compared to that in stage I/II (Figure 1C, P < 0.001). In addition, the high expression and low expression of DLEU1 exhibited significant differences in World Health Organization (WHO) stage (Table 1, P < 0.01). Meanwhile, DLEU1 expression in EC cell lines (Ishikawa, HEC-1A RL95-2 and KLE) was dramatically increased in contrast to that in the hESC cells (Figure 1D, P < 0.01). The expression of DLEU1 in Ishikawa and HEC-1A cell lines was higher than that in both RL95-2 and KLE cell lines. Therefore, Ishikawa and HEC-1A cell lines were chosen to perform the following experiments.
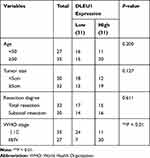 |
Table 1 Correlations Between lncRNA DLEU1 Expression and Clinicopathological Characteristics in EC |
LncRNA DLEU1 Knockdown Inhibits the Proliferation, Migration and Invasion of EC Cells in vitro and the Growth of Tumor Xenograft in vivo
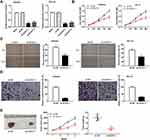
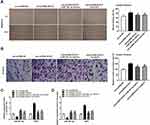
Following transfection of sh-DLEU1-1/-2 into Ishikawa and HEC-1A cells, the efficiency of transfection was detected by qRT-PCR. The results showed that DLEU1 expression was dramatically decreased after transfection of sh-DLEU1-1/-2 (Figure 2A, P < 0.01), which demonstrated that sh-DLEU1-1/-2 has been transfected into EC cells successfully. MTT assay showed that the viability of Ishikawa and HEC-1A cells was obviously reduced in the sh-DLEU1-1 group compared with the sh-NC group (Figure 2B, P < 0.01). Similarly, wound healing assay and transwell invasion assay, respectively, uncovered that the migration ability and invasion ability of EC cells were both restrained by DLEU1 knockdown (Figure 2C and D, P < 0.01). In addition, we also investigated the effect of DLEU1 knockdown on the growth of tumor xenograft in vivo. Four weeks later, the results demonstrated that in comparison to the sh-NC group, tumor volume and tumor weight were both distinctly decreased in the sh-DLEU1-1 group (Figure 2E, P < 0.01).
Exosomes are Uptaken by Ambient EC Cells
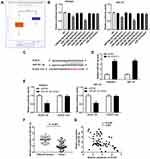
The exosomes isolated from Ishikawa cells were authenticated by Western blot assay. The results revealed that the expression levels of exosomal markers (CD63, CD9, CD81) in the exosomes group were increased obviously compared to those in the whole cells group (Figure 3A). Therefore, we confirmed that the exosomes were extracted successfully. After that, the fluorescence-labeled exosomes were observed using a confocal microscope, and we found that the exosomes were uptaken by Ishikawa receptor cells and intensively distributed around the nucleus (Figure 3B).
 |
Figure 3 Exosomes are uptaken by ambient EC cells. (A) Exosomes surface marker proteins CD63, CD9 and CD81 were detected by Western blot. (B) Uptake of exosomes was observed by a confocal microscope. |
LncRNA DLEU1 Targets miR-381-3p
To investigate the role of DLEU1 on miR-381-3p, we first detected miR-381-3p expression in UCEC and normal tissues in the TCGA database and found that miR-381-3p expression in UCEC tissues was decreased compared with the normal tissues (Figure 4A, P < 0.01). Based on Starbase database, eight miRNAs were chosen to screen out a target of DLEU1 by dual-luciferase reporter assay. As shown in Figure 4B, luciferase activity both in the miR-7-5p + DLEU1 group and miR-381-3p + DLEU1 group was significantly decreased in contrast to other groups (P < 0.01). Therefore, miR-381-3p was chosen to perform the subsequent trails due to the lower luciferase activity than miR-7-5p. Through Starbase software, potential binding site between DLEU1 and miR-381-3p was depicted (Figure 4C). To further identify whether DLEU1 directly binds to miR-381-3p, the expression of miR-381-3p was detected after transfection of sh-DLEU1-1. The results showed that DLEU1 knockdown promoted miR-381-3p expression (Figure 4D, P < 0.01). The luciferase activity in the DLEU1-WT/miR-381-3p mimics group was obviously reduced compared to the DLEU1-WT/miR-NC group (Figure 4E, P < 0.01). MiR-381-3p expression was down-regulated in tumor tissues in comparison to that in adjacent tissues (Figure 4F, P < 0.001). Besides, there was a significant inverse correlation between DLEU1 and miR-381-3p expression in EC tissues (Figure 4G, P < 0.001, r = −0.4397). The above data implied that miR-381-3p was a target of DLEU1 and was negatively modulated by DLEU1.
MiR-381-3p Inhibits the Proliferation, Migration, and Invasion of EC Cells
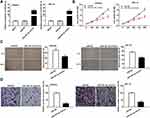
To investigate the role of miR-381-3p on the biological functions of EC cells, the transfection efficiency of miR-381-3p was detected. qRT-PCR results uncovered that miR-381-3p mimics significantly increased miR-381-3p expression (Figure 5A), suggesting that miR-381-3p mimics were successfully transfected into EC cells. Up-regulation of miR-381-3p suppressed the EC cell viability (Figure 5B, P < 0.01), and wound healing rate and number of invasion cells (Figure 5C and D, P < 0.01). These data indicated that up-regulation of miR-381-3p could inhibit the cell viability, migration ability and invasion ability of EC cells in vitro.
E2F3
To explore whether miR-381-3p regulates E2F3, we predicted the potential binding site between miR-381-3p and E2F3 using Targetscan software (Figure 6A). Dual-luciferase reporter assay displayed that the luciferase activity was dramatically reduced in the presence of wild type of E2F3 with miR-502-3p binding, but not miR-NC (Figure 6B). E2F3 expression was higher in tumor tissues than that in adjacent tissues (Figure 6C, P < 0.001) and was negatively correlated with miR-381-3p expression (Figure 6D, P < 0.001). miR-381-3p overexpression decreased E2F3 protein expression (Figure 6E). In a word, the above results demonstrated that E2F3 was the target gene of miR-381-3p and was negatively modulated by miR-381-3p.
Exosomal DLEU1 Accelerates the Migration and Invasion of EC Cells by Regulating miR-381-3p/E2F3 Axis
The effect of exosomal DLEU1 on the migration and invasion of EC cells was investigated using a co-culture model. The wound healing rate and number of invasion cells were both increased in the exo-pcDNA-DLEU1 group in contrast to the exo-pcDNA-NC group. However, up-regulation of miR-381-3p or down-regulation of E2F3 reversed the promoting effects of DLEU1 on the migration and invasion of Ishikawa cells (Figure 7A and B, P < 0.05). Subsequently, the expression of miR-381-3p and E2F3 in receptor cells and exosomes was detected. The results displayed that both in the receptor cells and exosomes, miR-381-3p expression was down-regulated in the exo-pcDNA-DLEU1 group, and the inhibitory role of DLEU1 overexpression on miR-381-3p was reversed by miR-381-3p mimics and E2F3 knockdown. Similarly, the promoting role of DLEU1 overexpression on E2F3 was also reversed by miR-381-3p overexpression and E2F3 knockdown (Figure 7C and D, P < 0.01). The obtained results implied that exosomal DLEU1 might promote the migration and invasion of EC cells by regulating the miR-381-3p/E2F3 axis in vitro.
Discussion
EC, the culprit of tumor-related death among women globally, is one of the most serious cancers of women.3 LncRNAs are taken part in the occurrence of several cancers, including EC.27,28 A recent study has displayed that the expression of PVT1 in EC tissues and cells is dramatically increased and PVT1 up-regulation appears closely related to tumor stage.27 LncRNA BANCR expression is also found to be markedly upregulated in EC tissues and is strongly associated with tumor stage.28 Similarly, this study revealed that DLEU1 expression was dramatically increased in EC tissues and cells and presented a notable correlation with tumor stage. Therefore, we thought that DLEU1 might be a pathogenic factor in EC.
LncRNAs have been found to act as important regulators in promoting the proliferation, migration and invasion, and inhibiting the apoptosis of EC cells.14,17 Previous studies reveal that CHL1-AS1 knockdown inhibits the viability and migration in EC cells,13 and silencing of NEAT116 or OGFRP129 suppresses the proliferation and invasion of EC cells. Similar to a previous study, down-regulation of DLEU1 can inhibit the proliferation, invasion, and migration of EC cells.17 In the current study, we found that DLEU1 inhibition suppressed the viability, migration ability and invasion ability of EC cells. Consequently, the results implied that DLEU1 might serve as a promoter in the occurrence and development of EC.
Increasing evidences indicate that miR-381 is participated in the progression of tumors30 and serves as a suppressor in several types of tumor.31–33 MiR-381 is found to be down-regulated in non-small cell lung cancer (NSCLC), and overexpression of miR-381 restrains the proliferation, migration and invasion of NSCLC cells.31 MiR-381 expression is obviously decreased in prostate cancer (PC) and breast cancer cells and miR-381 overexpression suppresses the invasion and proliferation of PC and breast cancer cells.32,33 In this study, we discovered that miR-381-3p expression was significantly decreased in EC tissues and cells. Furthermore, the proliferation, migration, and invasion of EC cells could be inhibited by miR-381-3p overexpression. In line with these results, Tu et al have identified that the expression of miR-381 is clearly reduced in EC tissues and cells and up-regulation of miR-381 inhibits the proliferation and invasion of EC cells.21 Moreover, miR-381-3p was a target of DLEU1 and negatively modulated by DLEU1, which implied that miR-381-3p might be regulated by DLEU1 to inhibit the progression of EC.
E2F3 is widely known as an oncogene to be involved in the development of several cancers.34,35 Up-regulation of E2F3 expression was observed in HCC tissues34 and in osteosarcoma (OS) tissues.35 In current study, we demonstrated that E2F3 expression in EC tissues was significantly increased. Consistently, Hu et al have showed that the expression of E2F3 is dramatically up-regulated in EC tissues.36 Meanwhile, E2F3 was proved to be a target gene of miR-381-3p and there was a notable inverse correlation between them. The results suggested that E2F3 might promote the progression of EC negatively regulated by miR-381-3p.
Abundance of reports have revealed that exosomes-mediated transfer of molecular changes micro-environment of tumor, eventually leading to carcinogenesis.11,37 ZFAS1 mediated by exosomes promotes the progress of esophageal squamous cell carcinoma (ESCC).38 Exosomal LNMAT2 induces tumorigenesis of bladder cancer.39 In this study, we revealed that exosomal DLEU1 overexpression increased the migration ability and invasion ability of EC cells. However, both up-regulation of miR-381-3p and down-regulation of E2F3 reversed the promoting effects of exosomal DLEU1 on the migration, and invasion in EC cells. Simultaneously, exo-pcDNA-DLEU1 down-regulated miR-381-3p expression and up-regulated E2F3 expression in both exosomes and receptor EC cells. The above results suggested that exosomal DLEU1 might promote the migration and invasion of EC cells by regulating the miR-381-3p/E2F3 axis.
Conclusions
In a word, this study uncovered exosomal DLEU1, which acts as an endogenous sponge of miR-381-3p to promote the proliferation, migration and invasion of EC cells. Overexpression of miR-381-3p down-regulates E2F3, inhibiting the progression of EC cells. The present study demonstrates that exosomal DLEU1/miR-381-3p/E2F3 axis are essential in EC progression, pointing to DLEU1 might act as a potential therapeutic target for EC.
Consent for Publication
Informed consent has been obtained for the publication of the data in this article.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethics Approval and Informed Consent
Human: the protocols of this study were reviewed and approved by ethical committee of The First Affiliated Hospital, Jinan University (Approval ID: 2019-LSPK-009). Each patient of this study obtained the written informed consent. Animal: The study was performed in the Animal Experimental Center of The First Affiliated Hospital, Jinan University. All animal experiments were undertaken according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and with the approval of ethical committee in The First Affiliated Hospital, Jinan University (Approval ID: 2018-LSPK-009).
Disclosure
The authors report no conflicts of interest for this work.
References
1. Oplawski M, Dziobek K, Zmarzly N, et al. Variances in the level of COX-2 and iNOS in different grades of endometrial cancer. Curr Pharm Biotechnol. 2020;21(1):52–59. doi:10.2174/1389201020666190918104105
2. Urick ME, Bell DW. Proteomic profiling of FBXW7-mutant serous endometrial cancer cells reveals upregulation of PADI2, a potential therapeutic target. Cancer Med. 2020;9(11):3863–3874. doi:10.1002/cam4.3013
3. Dahlgren E, Friberg L-G, Johansson S, et al. Endometrial carcinoma; ovarian dysfunction — a risk factor in young women. Eur J Obstet Gynecol Reprod Biol. 1991;41(2):143–150. doi:10.1016/0028-2243(91)90092-Y
4. Chen YL, Wang KL, Chen MY, et al. Risk factor analysis of coexisting endometrial carcinoma in patients with endometrial hyperplasia: a retrospective observational study of Taiwanese gynecologic oncology group. J Gynecol Oncol. 2013;24(1):14. doi:10.3802/jgo.2013.24.1.14
5. Wild PJ, Ikenberg K, Fuchs TJ, et al. p53 suppresses type II endometrial carcinomas in mice and governs endometrial tumour aggressiveness in humans. Der Pathologe. 2013;34(8):180. doi:10.1007/s00292-013-1859-x
6. Ce H, Tierney J, Symonds PR, Collingwood M, Green J. Chemotherapy for advanced, recurrent or metastatic endometrial carcinoma. Cochrane Database Syst Rev. 2005;5(4):CD003915.
7. Boll D, Verhoeven RHA, Van DA, et al. Incidence and survival trends of uncommon corpus uteri malignancies in the Netherlands, 1989–2008. Int J Gynecol Cancer. 2012;22(4):599–606. doi:10.1097/IGC.0b013e318244cedc
8. Simons M, Raposo G. Exosomes – vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21(4):575–581. doi:10.1016/j.ceb.2009.03.007
9. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi:10.1083/jcb.201211138
10. Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569. doi:10.1038/nri855
11. Hardin H, Helein H, Meyer K, Robertson S, Lloyd RV. Thyroid cancer stem-like cell exosomes: regulation of EMT via transfer of LncRNAs. Lab Investig. 2018;98(9):1133–1142. doi:10.1038/s41374-018-0065-0
12. Raghu K. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–1215. doi:10.1172/JCI81135
13. Shi Y, Zha J, Zuo M, Yan Q, Song H. Long noncoding RNA CHL1-AS1 promotes cell proliferation and migration by sponging miR-6076 to regulate CHL1 expression in endometrial cancer. J Cell Biochem. 2020;121(3):2655–2663. doi:10.1002/jcb.29486
14. Zhang Y, Zhang P, Chen L, Zhao L, Zhu J, Zhu T. The long non-coding RNA-14327.1 promotes migration and invasion potential of endometrial carcinoma cells by stabilizing the potassium channel Kca3.1. Onco Targets Ther. 2019;12:10287–10297. doi:10.2147/OTT.S226737
15. Treeck O, Skrzypczak M, Schuler-Toprak S, Weber F, Ortmann O. Long non-coding RNA CCAT1 is overexpressed in endometrial cancer and regulates growth and transcriptome of endometrial adenocarcinoma cells. Int J Biochem Cell Biol. 2020;122:105740. doi:10.1016/j.biocel.2020.105740
16. Dong P, Xiong Y, Yue J, et al. Long noncoding RNA NEAT1 drives aggressive endometrial cancer progression via miR-361-regulated networks involving STAT3 and tumor microenvironment-related genes. J Exp Clin Cancer Res. 2019;38(1):295. doi:10.1186/s13046-019-1306-9
17. Du Y, Wang L, Chen S, Liu Y, Zhao Y. lncRNA DLEU1 contributes to tumourigenesis and development of endometrial carcinoma by targeting mTOR. Mol Carcinog. 2018;57(9):1191–1200. doi:10.1002/mc.22835
18. Shao W, Li Y, Chen F, Jia H, Jia J, Fu Y. Long non-coding RNA DLEU1 contributes to the development of endometrial cancer by sponging miR-490 to regulate SP1 expression. Die Pharmazie. 2018;73(7):379–385.
19. Tsuruta T, Kozaki KI, Uesugi A, et al. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res. 2011;71(20):6450–6462. doi:10.1158/0008-5472.CAN-11-0364
20. Le XF, Merchant O, Bast RC, Calin GA. The roles of microRNAs in the cancer invasion-metastasis cascade. Cancer Microenvironment. 2010;3(1):137–147. doi:10.1007/s12307-010-0037-4
21. Tu C, Wang F, Wan J. MicroRNA-381 inhibits cell proliferation and invasion in endometrial carcinoma by targeting the IGF-1R. Mol Med Rep. 2017. doi:10.3892/mmr.2017.8288
22. Oeggerli M, Tomovska S, Schraml P, et al. E2F3 amplification and overexpression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Oncogene. 2004;23(33):5616–5623. doi:10.1038/sj.onc.1207749
23. Hua M, Qin Y, Sheng M, et al. E2F3. Mol Med Rep. 2019;19(5):3575–3583.
24. Guo J, Zhang J, Yang T, Zhang W, Liu M. MiR-22 suppresses the growth and metastasis of bladder cancer cells by targeting E2F3. Int J Clin Exp Pathol. 2020;13(3):587–596.
25. Pei Y, Tang Z, Cai M, Yao Q, Xie B, Zhang X. The E2F3/miR-125a/DKK3 regulatory axis promotes the development and progression of gastric cancer. Cancer Cell International. 2019;19(1). doi:10.1186/s12935-019-0930-y
26. Wan J, Liu H, Feng Q, Liu J, Ming L. HOXB9 promotes endometrial cancer progression by targeting E2F3. Cell Death Dis. 2018;9(5):509. doi:10.1038/s41419-018-0556-3
27. Kong F, Ma J, Yang H, Yang D, Wang C, Ma X. Long non-coding RNA PVT1 promotes malignancy in human endometrial carcinoma cells through negative regulation of miR-195-5p. Biochim Biophys Acta Mol Cell Res. 2018;1865(10):1479–1490. doi:10.1016/j.bbamcr.2018.07.008
28. Wang D, Wang D, Wang N, Long Z, Ren X. Long non-coding RNA BANCR promotes endometrial cancer cell proliferation and invasion by regulating MMP2 and MMP1 via ERK/MAPK signaling pathway. Cell Physiol Biochem. 2016;40(3–4):644–656. doi:10.1159/000452577
29. Lv Y, Chen S, Wu J, et al. Upregulation of long non-coding RNA OGFRP1 facilitates endometrial cancer by regulating miR-124-3p/SIRT1 axis and by activating PI3K/AKT/GSK-3beta pathway. Artif Cells Nanomed Biotechnol. 2019;47(1):2083–2090. doi:10.1080/21691401.2019.1617727
30. Liu C, Tian X, Zhang J, Jiang L. Long non-coding RNA DLEU1 promotes proliferation and invasion by interacting with miR-381 and enhancing HOXA13 expression in cervical cancer. Front Genet. 2018;9:629.
31. Jin D, Guo J, Wu Y, Chen W, Su G. Metformin-repressed miR-381-YAP-snail axis activity disrupts NSCLC growth and metastasis. J Exp Clin Cancer Res. 2020;39(1). doi:10.1186/s13046-019-1503-6
32. Rui X, Gu T, Pan H, Shao S, Shao H, Wang J. MicroRNA‑381 suppresses proliferation and invasion of prostate cancer cells through downregulation of the androgen receptor. Oncol Lett. 2019;18(2):2568–2575. doi:10.3892/ol.2019.10539
33. Pour ZB, Nourbakhsh M, Mousavizadeh K, Madjd Z, Mobaser SE. Up-regulation of MIR-381 inhibits NAD + salvage pathway and promotes apoptosis in breast cancer cells. Excli J. 2019;683–696.
34. Cao T, Li H, Hu Y, Ma D, Cai X. miR-144 suppresses the proliferation and metastasis of hepatocellular carcinoma by targeting E2F3. Tumor Biol. 2014;35(11):10759–10764. doi:10.1007/s13277-014-2017-7
35. Dong D, Gong Y, Zhang D, Bao H, Gu G. miR-874 suppresses the proliferation and metastasis of osteosarcoma by targeting E2F3. Tumor Biol. 2015;37(5):1229–1236. doi:10.1007/s13277-015-3867-3
36. Hu J, Shen J, Sun J. CDK4/RB/E2Fs axis as potential therapeutic target of endometrial cancer. Biomed Pharmacother. 2020;125:109870. doi:10.1016/j.biopha.2020.109870
37. Li L, Li C, Wang S, et al. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to Normoxic cells to elicit a prometastatic phenotype. cancer res. 2016;
38. Li Z, Qin X, Bian W, et al. Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA-124/STAT3 axis. J Exp Clin Cancer Res. 2019;38(1):477. doi:10.1186/s13046-019-1473-8
39. Chen C, Luo Y, He W, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest. 2020;130(1):404–421. doi:10.1172/JCI130892
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.