Back to Journals » OncoTargets and Therapy » Volume 14
Exonuclease 1 (EXO1) is a Potential Prognostic Biomarker and Correlates with Immune Infiltrates in Lung Adenocarcinoma
Authors Zhou CS , Feng MT, Chen X, Gao Y, Chen L, Li LD, Li DH, Cao YQ
Received 21 October 2020
Accepted for publication 16 January 2021
Published 15 February 2021 Volume 2021:14 Pages 1033—1048
DOI https://doi.org/10.2147/OTT.S286274
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Gaetano Romano
Chang-shuai Zhou,1,2,* Ming-tao Feng,1,2,* Xin Chen,1,2 Yang Gao,1,2 Lei Chen,1,2 Liang-dong Li,1,2 De-heng Li,1,2 Yi-qun Cao1,2
1Department of Neurosurgery, Fudan University Shanghai Cancer Center, Shanghai, People’s Republic of China; 2Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yi-qun Cao Email [email protected]
Background: Exonuclease 1 (EXO1) has been identified to be highly expressed in different human malignancies, but its expression and prognostic role in lung adenocarcinoma (LUAD) remain unknown.
Materials and Methods: Two independent cohorts extracted from public databases and one cohort from our center were analyzed in this study. Expression levels of EXO1 in LUAD tissues and paired para-cancer tissues were detected. The prognostic value of EXO1 in LUAD patients was evaluated in the three cohorts. Enrichment analyses were performed to explore the possible underlying biological pathways. Moreover, we also explored the correlations between EXO1 and tumor-infiltrating immune cells and evaluated the impact of EXO1 knock-down on the migration of lung cancer cells.
Results: In this study, we found that EXO1 was highly expressed in LUAD tissues compared with para-cancerous tissues in public databases (p < 0.01), which was consistent with our data (p < 0.01). Survival analysis indicated that high expression of EXO1 was associated with poor prognosis in LUAD (p < 0.01). Enrichment analyses indicated that biological pathways like cell cycle regulation, DNA damage and repair, immune response, neuroactive ligand-receptor interaction, may be associated with EXO1 aberrant expression. Moreover, high expression of EXO1 was correlated with decreased infiltrating B cells (p < 0.01) and CD4+ T cells (p < 0.01) levels, and low infiltrating levels of B cells (p < 0.01) and dendritic cells (DCs) (p < 0.05) indicated poor overall survival (OS) in LUAD. Additionally, in vitro experiments suggested that knockdown of EXO1 may inhibit the migratory ability of lung cancer cells.
Conclusion: In conclusion, EXO1 is a potential prognostic biomarker in LUAD, and correlates with infiltrating levels of immune cells in the tumor microenvironment. Further prospective validation of EXO1 in lung cancer is warranted.
Keywords: EXO1, lung adenocarcinoma, prognosis, biomarker, immune infiltration
Introduction
Lung cancer (LC) is one of the most common malignancies and is a leading cause of cancer-associated mortality among different cancers.1,2 Histologically, LC can be divided into two subsets: small cell lung cancer and non-small cell lung cancer (NSCLC). In addition, LUAD accounts for the largest subgroup of NSCLC and exhibits a tendency for metastasis at an early stage.3,4 Recently, novel therapies for LC have been created to exploit the complex interplay of several treatments including surgery, systemic chemotherapy, radiation therapy, targeted therapy, and immunotherapy.5–7 However, the recurrence and mortality rates of LUAD are still high, and the prognosis of patients is poor.8,9 Considering the low survival rate and poor status of LUAD patients, it is imperative to find more effective biomarkers to facilitate novel therapeutic methods.
As a 5ʹ to 3ʹ exonuclease, EXO1 was found to play vital roles in DNA mismatch repair (MMR), DNA double-strand break repair (DSBR), cell cycle regulation, nucleotide excision repair, immunoglobulin maturation, and telomere maintenance.10–13 Furthermore, EXO1 can mediate the MMR process by interacting with MSH2 or MSH2-MSH6 form heterodimers.14,15 Dysfunctional DNA replication and mismatch repair may lead to the initiation and progression of tumors. Accordingly, it has been reported that increased EXO1 expression is associated with tumor invasion, metastasis, and poor OS in different tumors. For instance, a high expression level of EXO1 is correlated with larger tumor size, increased lymph node metastasis, and poor overall survival in hepatocellular carcinoma.16 Similarly, the overexpression of EXO1 indicates poor survival of prostate cancer patients and promotes tumor progression and metastasis.17 Moreover, a high expression level of the EXO1 protein is associated with poor OS in breast cancer patients.18 In contrast, it has also been revealed that high EXO1 expression favorably affects OS in colorectal cancer patients.19 Studies examining the contributions of several EXO1 single-nucleotide polymorphisms (SNPs) to lung cancer risk have been reported,20–22 but few studies were conducted to explore the prognostic value of EXO1 levels in LUAD patients.
In this study, we analyzed the EXO1 expression levels in tumor and adjacent normal tissues and explored the prognostic value of EXO1 in LUAD patients. To investigate the underlying mechanisms, we performed enrichment analyses and evaluated the associations between EXO1 expression and tumor-infiltrating immune cells. Furthermore, in vitro experiments were performed to evaluate the impact of EXO1 expression on lung cancer cells. Above all, we identified a previously unconfirmed potential biomarker for LUAD patients and explored the possible mechanisms.
Materials and Methods
Patients and Cell Lines
Cohort 1 was extracted from the Cancer Genome Atlas (TCGA) database, including RNA-seq data generated from 483 LUAD tissues and 59 normal tissues. Cohort 2, GSE31210, was obtained from the Gene Expression Omnibus (GEO) database, containing expression profiles of 226 lung adenocarcinomas and 20 normal tissues. A cohort of 78 LUAD patients from Fudan University Shanghai Cancer Center, including formalin-fixed paraffin-embedded carcinomas and the adjacent normal tissues, was also collected. Two LUAD cell lines (A549 and H1299) used in this study were obtained from the Chinese Academy of Science (Shanghai, China).
Expression and Survival Analysis
We overviewed the genetic alterations of EXO1 in LUAD tissues by using the cBioPortal platform (www.cbioportal.org).23 Then, the mRNA levels of EXO1 in human pan-cancer tissues and normal tissues were also explored using the ONCOMINE database (www.oncomine.org).24 Next, we estimated the associations between EXO1 expression with its promoter methylation status, TP53 mutation status, and clinicopathological features via the UALCAN database (http://ualcan.path.uab.edu).25 The prognostic value of EXO1 mRNA expression in cohort 1 was analyzed through the GEPIA database (http://gepia.cancer-pku.cn/).26 The differential expression of EXO1 in tumor tissues and para-carcinoma tissues and its prognostic value in LUAD were verified in cohort 2 from the GEO database. Correlation analysis of Ki-67 with EXO1 was conducted in LUAD from TCGA through GPEPA.
Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis and Gene Set Enrichment Analysis (GSEA)
Base on the gene expression levels of EXO1, differentially expressed genes (DEGs) were retrieved from samples from the TCGA database by using the limma R package. The thresholds for DEGs were |log2FC| ≥ 1 and adjusted p < 0.05. KEGG analysis was carried out according to the filtering criteria mentioned above (http://www.genome.jp/kegg/pathway.html). Also, GSEA was applied to detect functional categories enriched in upregulated DEGs (http://software.broadinstitute.org/gsea/index.jsp). The normalized enrichment score (NES) was recorded, and a false discovery rate (FDR) < 0.05 was set as the threshold.
Correlations with Immune Infiltrating Cells
The infiltration levels of six immune cell types, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages and DCs, associated with EXO1 expression were analyzed via the Tumor Immune Estimation Resource (TIMER) database (https://cistrome.shinyapps.io/timer/).27 Then, survival analyses of tumor-infiltrating immune cells in LUAD patients were conducted with Kaplan-Meier method, and comparisons of survival curves were analyzed with Log rank test.
Protein-Protein Interaction (PPI) Network Analysis
PPI network analysis was performed to analyze the correlation among the DEGs via Search Tool for the Retrieval of Interacting Genes (STRING) (http://string-db.org/), and the top 25 relevant hub genes with the maximum specific weigh modules were shown.28 The functions of these hub genes were described in detail according to the Gene Cards description (https://www.genecards.org/).29
Immunohistochemistry
Primary antibody specific for EXO1 (Affinity, Df3615, 1:100) was incubated with tissue sections at 4°C overnight. Next, incubation with a secondary goat anti-rabbit HRP-conjugated antibody (Servicebio, GB23303, 1:200) was performed for 50 minutes. Then, the sections underwent diaminobenzidine color development. The sections were counterstained with hematoxylin after being washed with water and phosphate buffer saline. Finally, microscopic examination, image acquisition and analysis were conducted. Counting was performed using the H-Score algorithm. H-score = (percentage of cells with weak intensity × 1) + (percentage of cells with moderate intensity × 2) + (percentage of cells with strong intensity × 3).30 Correlation analysis of Ki-67 index and EXO1 H-score was performed in our cohort.
Cell Transfection
Small interfering RNA (siRNA) against EXO1 and corresponding negative control (si-NC) were constructed by Genepharma (Shanghai, China). The siRNA sequences were subjected to BLAST to reduce off-target effects. The constructed siRNA sequences and si-NC sequences were transferred into cells using Lipofectamine 2000. The siRNA sequences were as follows: si-EXO1-1: 5′-CCUCUUUGCCUGAGAAUAATT-3′, and si-EXO1-2: 5′-GCGGAAGAGGAUAUAUUUATT-3′. The effect of the EXO1 knockdown was assessed by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) 48 h after the transfection.
qRT-PCR Assays
Total RNA from the lung cancer cells was extracted using TRIzo006C reagent (Invitrogen, CA, USA), and 1 µg RNA was reverse transcribed into cDNA by using the reverse transcriptase M-MLV (Invitrogen, CA, USA). QRT-PCR was performed by using SYBR-Green PCR master mix (Invitrogen, CA, USA) on a Roche Light Cycler 480 system (Roche Diagnostics, Mannheim, Germany). β-actin was used as an endogenous control. The relative expression of genes was calculated using the formula 2-∆Ct (∆Ct = Ct target - Ct control). The gene primer sequences were as follows: β-actin forward 5′-CCCTGGCTCCTAGCACCAT-3′, β-actin reverse 5′-GATAGAGCCACCAATCCACACA-3′, EXO1 forward, 5ʹ- AGGACCATTTCACCACCCACTTTG-3ʹ, and EXO1 reverse, 5ʹ- AGGACCATTTCACCACCCACTTTG-3ʹ.
Transwell Migration Assays
For cell migration assays, EXO1-negative control and EXO1-knockdown lung cancer cells (A549 and H1299) were added to the upper chambers in Dulbecco’s modified eagle medium (DMEM) containing 1% fetal bovine serum (FBS). The lower chambers were filled with DMEM containing 20% FBS. After a 24-hour incubation, the upper chambers were fixed with methanol at room temperature for 30 minutes and stained with crystal violet staining solution for an additional 30 minutes. The cells that passed through the membrane were counted under a Leica microscope (magnification, ×100).
Statistical Analysis
Statistical analyses were performed using SPSS, version 22 (IBM, NY, USA) and GraphPad Prism 7 software (San Diego, CA, USA). Survival analysis was performed by Kaplan-Meier survival analysis, using the median or quartile value as a cutoff. Then, the prognostic value of each potential risk factor was analyzed by the Cox proportional hazard regression model, and the estimated hazard ratio (HR) and associated 95% confidence interval (CI) were demonstrated. A two-sided p < 0.05 was considered statistically significant. Student’s t-test was used for comparing parameters between groups. Two levels of significance for comparisons between groups (*p < 0.05, **p < 0.01) were used.
Results
Elevated mRNA Expression of EXO1 in LUAD
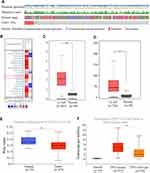
Although mutations were infrequent in tested patients, increased amplification was most frequently observed among EXO1 genetic alterations (p<0.001) (Figure 1A and S1). Then, we found a higher expression of EXO1 in LUAD patient samples than that in normal tissue samples in the TCGA database (p < 0.01) (Figure 1C), which was confirmed in the GSE31210 dataset (Figure 1D). Next, the promoter methylation level of EXO1 was lower in LUAD tissues compared with normal tissues (p < 0.05) (Figure 1E). Our findings also indicated that EXO1 expression was significantly higher in the TP53-mutant group than that in the TP53-wild-type group (p < 0.05) (Figure 1F). Furthermore, we observed that EXO1 was overexpressed in different cancers compared with normal tissues from the Oncomine database (Figure 1B).
High EXO1 Level Correlated with Clinicopathological Features and Indicated Poor OS in LUAD Patients
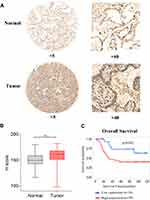
The overexpression of EXO1 in LUAD was significantly associated with a more advanced tumor stage (p < 0.05) (Figure 2A), lymph node metastasis (p < 0.05) (Figure 2B) and distant metastasis (p < 0.05) (Figure 2C). Kaplan-Meier survival analysis showed that patients with higher expression of EXO1 mRNA had poorer disease-free survival (DFS) (p < 0.01, Figure 2D) and OS (p < 0.01, Figure 2E). Similar results were also observed based on survival analysis from the GSE31210 dataset (p < 0.001, Figure 2F). Both univariate and multivariate Cox regression analysis indicated that a high level of EXO1 was correlated with poor OS among the patients from the TCGA database (p < 0.001) (Table 1). Moreover, clinicopathological features like advanced T stage (p < 0.01) and metastatic lymph node (p < 0.001) were also associated with poor OS in LUAD (Table 1). A cohort of 78 paired human LUAD and para-cancerous tissue samples from our center were also analyzed by immunohistochemistry. Higher protein expression of EXO1 was observed in tumor tissues compared with that in paired adjacent tissues (p < 0.001) (Figure 3A and B). Kaplan-Meier survival analysis suggested that a higher EXO1 immunohistochemistry score suggested a poorer OS in LUAD patients from our center (p < 0.05) (Figure 3C). The univariate and multivariate regression Cox analyses were also conducted, and the results indicated that upregulation of EXO1 was associated with poor OS in LUAD (p < 0.05) (Table 2). Furthermore, older age (p < 0.01) and advanced T stage (p < 0.001) were also correlated with poor OS in LUAD patients (Table 2). In addition, EXO1 expression was significantly correlated with Ki-67 in LUAD based on TCGA (R=0.6, p=0.00), as shown in Figure S2. And yet the positive correlation between EXO1 H-score and Ki-67 index in our cohort was not significant (R=0.11, p=0.34), as shown in Figure S3. Baseline features of included patients were listed in Supplementary Tables (Table S1, S2).
 |
Table 1 Cox Regression Analysis for Overall Survival of LUAD Patients from the TCGA Database |
 |
Table 2 Cox Regression Analysis for Overall Survival of 78 LUAD Patients in Our Center |
Enrichment Analysis of the EXO1 Expression Phenotype
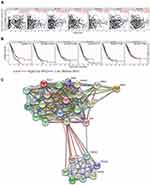
After analyzing the gene expression profiles of LUAD samples in the TCGA database, a total of 2262 DEGs were identified between samples with high or low expression of EXO1. Of these DEGs, 732 genes exhibited downregulated expression, and 1530 genes exhibited upregulated expression, as shown in the volcano plot (Figure 4A). The top 10 enriched signaling pathways for the DEGs were determined by KEGG analysis (Figure 4B). We also performed GSEA of the upregulated DEGs in LUAD, and the top 10 enriched biological pathways were illustrated (Figure 4C). Enrichment analyses disclosed several enriched signaling pathways, which were mainly involved in cell cycle regulation (cell cycle, DNA replication, oocyte meiosis), DNA damage and repair (nicotine addiction, alcoholism, MMR, nucleotide excision repair, homologous recombination), immune response (systemic lupus erythematosus, complement and coagulation cascades), and neuroactive ligand-receptor interaction. Common pathways identified by both GSEA and KEGG analyses were attributed to three pathways, namely, cell cycle (Figure 4D), oocyte meiosis (Figure 4E), and DNA replication (Figure 4F).
EXO1 Correlated with Immune Infiltration Levels and May Affect the Outcome in LUAD Patients
Negative correlations between EXO1 expression and the levels of infiltrating B cells (r = − 0.219, p < 0.01) and CD4+ T cells (r = − 0.124, p < 0.01) in LUAD were shown in Figure 5A. Additionally, cumulative survival analysis indicated that low levels of infiltrating B cells (p < 0.01) and DCs (p < 0.05) might predict poor OS in LUAD patients (Figure 5B).
PPI Network Interacted with EXO1
To explore the potential functional connections of DEGs, a PPI network was constructed using the STRING database. The top 25 genes with the most significant degree of connectivity with EXO1 were shown in Figure 5C, and the names, abbreviations, and functions of these genes were listed in Table 3. The functions of hub genes mainly were concentrated in biological pathways such as cell cycle regulation (PCNA, SFN, YWHA family members), MMR (MLH1, PMS1, PMS2, MSH2, MSH3), DSBR (MRE11A, RAD50, RAD52, UBE2I), immune response (ATM, XRCC5, XRCC6).
 |
Table 3 Top 25 Hub Genes in PPI Network of EXO1 |
Knockdown of EXO1 Inhibited the Migration of Lung Cancer Cells in vitro
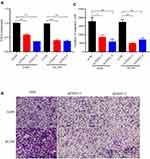
Transwell migration experiments were conducted to explore the impact of EXO1 expression levels on lung cancer cells. Our results revealed that knockdown of EXO1 expression significantly reduced the migration numbers of A549 and H1299 cells (Figure 6A–C). These findings indicated that knockdown of EXO1 may reduce the migratory ability of lung cancer cells in vitro experiments.
Discussion
Despite advances in lung cancer therapy, this disease is still among the most malignant types of cancers with a poor 5-year survival rate.2,5,31 Therefore, the identification of new effective biomarkers and exploration of the underlying molecular mechanisms are urgently needed. Persistent DNA damage and dysfunction of the DNA damage repair process may induce genomic instability, leading to cancer initiation, progression, and metastasis.32,33 However, degradation-resistant EXO1 could result in DNA hyper-resection in homologous recombination, which severely interfered with DSBR and induced chromosomal instability.34 Additionally, several EXO1 SNPs have been reported to be associated with increased susceptibility to lung cancer, especially in patients with a smoking history.20,21,35 Further bioinformatic analysis study showed that EXO1 was one of the hub genes acting as biomarkers of poor prognosis in lung cancer.36 Here, our study revealed that high EXO1 expression was consistently correlated with poor prognosis in LUAD, as well as with advanced malignant stage, lymphatic metastasis, and distant metastasis, via bioinformatic analysis. Next, the higher expression of EXO1 in LUAD compared with para-cancerous tissues was confirmed with immunohistochemistry in our cohort. Combined with genetic alterations of EXO1, both promoter hypomethylation and gene amplification may account for the increased EXO1 expression in LUAD (Figure 1A and S1). Furthermore, patients with high EXO1 protein expression had a poorer prognosis when the cutoff value was set at quartile. All these findings suggested a potential oncogenic role for EXO1 in LUAD, which was consistent with previous studies of EXO1 in lung cancer36 and other cancers, such as hepatocellular carcinoma,16 prostate cancer,17 colorectal adenoma,37 and astrocytoma.38
To further explore the underlying mechanisms, we performed enrichment analyses of DEGs according to EXO1 expression levels. The results demonstrated that the enriched pathways were mainly involved in cell cycle regulation, DNA damage repair, and immune response, which was consistent with the functions of hub genes of the PPI network. Accordingly, it has been reported that EOX1 colocalizes with the cell cycle regulatory protein proliferating cell nuclear antigen (PCNA) in the S phase,39 which further stimulates EXO1 to perform DNA end resection process.40 Moreover, cell cycle-associated proteins have been shown to modulate the effectiveness of radiation and chemotherapy.41,42 Several SNPs of EXO1 have been confirmed to be associated with radiotherapy or chemotherapy resistance, such as radiotherapy resistance in glioblastoma cell lines,38 chemotherapy resistance in ovarian cancer,43 and resistance to cisplatin treatment in NSCLC patients.44 And in our study, we found that EXO1 was highly expressed in TP53-mutant group compared with TP53-wild-type group in LUAD. To eliminate the confounding effect by TP53 status, multivariate Cox analysis were conducted. As shown in Table 1, TP53 status was not an independent prognostic factor in cohort 1. Furthermore, our findings showed that EXO1 was significantly and positively correlated with Ki-67 expression in the TCGA cohort (Figure S2). However, perhaps due to our small sample size, the positive correlation between EXO1 H-score and Ki-67 index was not significant in our cohort (Figure S3). These results may be explained by the potentially important roles of EXO1 in cell cycle regulation, which could further promote tumor cell proliferation and may affect the sensitivity to chemotherapy and radiation in LUAD patients.
Previous studies suggested that genomic instability which was caused by dysfunctional DNA damage repair may lead to tumor initiation, invasion, metastasis, and immune evasion.45–48 As mentioned above, genomic instability may be caused by aberrant expression of EXO1, involving abnormalities in DNA mismatch repair,49 telomere maintenance,12 and homologous recombination.34,50 In our research, we found that high expression of EXO1 was correlated with lymph node metastasis and distant metastasis in data from public databases. Additionally, previous studies suggested that DNA damaging factors like smoking history and nicotine consumption could suppress innate immune function and promote lung cancer brain metastasis.51,52 However, our results showed no difference in the proportion of distant metastasis between high and low EXO1 expression groups in our cohort, possibly due to the small size of cases. Interestingly, as an outstanding enriched pathway in KEGG, neuroactive ligand-receptor interaction was reported to be closely related to smoking-induced lung cancers.53 Importantly, it has been found that neuroactive ligand-receptor interaction might play important roles in brain metastasis of triple-negative breast cancer.54 Our data indicated that knockdown of EXO1 expression significantly reduces the migratory capacity of lung cancer cells in vitro. These findings may deliver a hint for the role of EXO1 in distant metastasis of lung cancer, which requires further investigation.
Tumor-infiltrating immune cells, such as tumor-associated macrophages (TAMs), CD8+ T cells, CD4+ T cells, B cells, are important components of the lung cancer microenvironment.55–58 To explore the possible roles of EXO1 in the immune system, we analyzed the tumor-infiltrating immune cell levels according to EXO1 expression through the TIMER database. Decreased TAMs and increased CD8+ T infiltration levels in LUAD were protective factors in previous studies.55,57 In line with previous studies, we also found that the number of macrophages in tumor tissues was more and the staining of EXO1 on macrophages was deeper than that in normal tissues (Figure 3A). However, our findings showed that high EXO1 expression level was significantly correlated with decreased infiltrating B cells and CD4+ T cells levels, but not with CD8+ T cells. Accordingly, EXO1 played vital roles in immunoglobulin maturation and antibody diversity through class switching recombination and somatic hypermutation.59,60 Therefore, aberrant overexpression of EXO1 may result in the dysfunction of immunoglobulin maturation and reduction of antibody diversity, which may further decrease the infiltrating B cell levels.34 Furthermore, B cell depletion impaired CD4+ T cell activation and clonal expansion in mice whereas CD8+ T cell activation was not affected.61 Meanwhile, CD4+ T cell receptor repertoire clonality might be downregulated by decreasing the density of B cells in lung cancer.62 We suggest that EXO1 may induce an immune-suppressive environment for LUAD patients by decreasing B cells and CD4+ T cell infiltration, but not CD8+ T cells. Moreover, the abundance of immune cells is varied in different immune subtypes. For instance, the Immunity High subtype generally has high infiltrating levels of CD8+ T cells and exhibits more sensitivity to immunotherapy.63 In contrast, the Immunity Low subtype tends to obtain a high capacity of immune evasion for neoantigen loss and low infiltrating levels of CD8 + T cells.64 As mentioned above, we speculate that the humoral immunity inactivation caused by EXO1 aberrant overexpression may induce a tumor environment similar to the Immunity Low subtype. In the immune-suppressive environment, CD8+ T cell infiltrating levels may not have a significantly impact on the outcome in LUAD patients who are insensitive to immunotherapy.63 Importantly, our results also revealed that low infiltrating levels of B cells were significantly associated with poor OS in LUAD patients. Consistently, a high density of tumor-associated tertiary lymphoid structure B cells was confirmed as a biomarker of favorable overall survival in NSCLC.65 Future studies are needed to validate our hypothesis and extend our findings.
However, there were still some limitations in this study. First, the small sample size of our own cohort and retrospective design might have led to patient selection bias. Therefore, further large sample size studies are needed to verify our findings. Second, we merely performed the migration assay in vitro, and other in vitro experiments as well as in vivo experiments will be conducted in future research. Finally, the relationships between EXO1 expression levels and immune cells were analyzed from data deposited in public databases. Prospective studies are needed to further validate the potential roles of EXO1 in the immune microenvironment of LUAD patients.
Conclusion
In summary, our study suggested that EXO1 was a potential marker to predict the poor prognosis of LUAD patients, and correlated with tumor-infiltrating immune cells. Further research is needed to confirm these findings.
Abbreviations
CI, confidence interval; DCs, dendritic cells; DEGs, differentially expressed genes; DFS, disease-free survival; DMEM, Dulbecco’s modified eagle medium; DSBR, DNA double-strand break repair; EXO1, Exonuclease 1; FBS, fetal bovine serum; FDR, False Discovery Rate; GEO, Gene Expression Omnibus; GSEA, gene set enrichment analysis; HR, hazard ratio; KEGG, Kyoto Encyclopedia of Genes and Genomes; LC, lung cancer; LUAD, lung adenocarcinoma; MMR, DNA mismatch repair; NES, normalized enrichment score; NSCLC, non-small cell lung cancer; OS, overall survival; PCNA, protein proliferating cell nuclear antigen; PPI, protein-protein interaction; qRT-PCR, quantitative reverse tran-scription-polymerase chain reaction; SNPs, single-nucleotide polymorphisms; STRING, Search Tool for the Retrieval of Interacting Genes; TAMs, tumor-associated macrophages; TCGA, the Cancer Genome Atlas; siRNA, small interfering RNA.
Ethical Approval
This study was approved by the Ethical Committee of Fudan University Shanghai Cancer Center and was conducted following the Declaration of Helsinki. Written informed consent was obtained from all patients or their authorized family members (if the participant was incompetent in writing their name due to low education). The purpose of the study was confirmed to be fully explained and understood by all patients.
Funding
This work was supported by funds from the Natural Science Foundation of China (Grant No.81802494) and Shanghai Municipal Commission of Health and Family Planning (Grant No. 20184Y0107).
Disclosure
No potential conflicts of interest were disclosed.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69(5):363–385. doi:10.3322/caac.21565
3. Patel MI, Cheng I, Gomez SL. US lung cancer trends by histologic type. Cancer. 2015;121(7):1150–1152. doi:10.1002/cncr.29180
4. Kim YH, Nagai H, Ozasa H, et al. Therapeutic strategy for non-small-cell lung cancer patients with brain metastases (Review). Biomed Rep. 2013;1(5):691–696. doi:10.3892/br.2013.151
5. Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol. 2019;16(6):341–355. doi:10.1038/s41571-019-0173-9
6. Moya-Horno I, Viteri S, Karachaliou N, et al. Combination of immunotherapy with targeted therapies in advanced non-small cell lung cancer (NSCLC). Ther Adv Med Oncol. 2018;10:10. doi:10.1177/1758834017745012
7. Osmani L, Askin F, Gabrielson E, et al. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol. 2018;52:103–109. doi:10.1016/j.semcancer.2017.11.019
8. Forde PM, Chaft JE, Smith KN, et al. Neoadjuvant PD-1 blockade in resectable lung Cancer. N Engl J Med. 2018;378(21):1976–1986. doi:10.1056/NEJMoa1716078
9. Pikor LA, Ramnarine VR, Lam S, et al. Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung Cancer. 2013;82(2):179–189. doi:10.1016/j.lungcan.2013.07.025
10. Keijzers G, Liu D, Rasmussen LJ. Exonuclease 1 and its versatile roles in DNA repair. Crit Rev Biochem Mol Biol. 2016;51(6):440–451. doi:10.1080/10409238.2016.1215407
11. Keijzers G, Bakula D, Petr MA, et al. Human exonuclease 1 (EXO1) regulatory functions in DNA replication with putative roles in cancer. Int J Mol Sci. 2018;20(1):20. doi:10.3390/ijms20010074
12. Xue Y, Marvin ME, Ivanova IG, et al. Rif1 and Exo1 regulate the genomic instability following telomere losses. Aging Cell. 2016;15(3):553–562. doi:10.1111/acel.12466
13. Goellner EM, Putnam CD, Kolodner RD. Exonuclease 1-dependent and independent mismatch repair. DNA Repair (Amst). 2015;32:24–32. doi:10.1016/j.dnarep.2015.04.010
14. Goellner EM, Putnam CD, Graham WJ, et al. Identification of Exo1-Msh2 interaction motifs in DNA mismatch repair and new Msh2-binding partners. Nat Struct Mol Biol. 2018;25(8):650. doi:10.1038/s41594-018-0092-y
15. Goellner EM, Smith CE, Campbell CS, et al. PCNA and Msh2-Msh6 activate an mlh1-pms1 endonuclease pathway required for Exo1-independent mismatch repair. Mol Cell. 2014;55(2):291–304. doi:10.1016/j.molcel.2014.04.034
16. Dai Y, Tang Z, Yang Z, et al. EXO1 overexpression is associated with poor prognosis of hepatocellular carcinoma patients. Cell Cycle. 2018;17(19–20):2386–2397. doi:10.1080/15384101.2018.1534511
17. Luo F, Wang Y-Z, Lin D, et al. Exonuclease 1 expression is associated with clinical progression, metastasis, and survival prognosis of prostate cancer. J Cell Biochem. 2019;120(7):11383–11389. doi:10.1002/jcb.28415
18. Qi L, Zhou B, Chen J, et al. Significant prognostic values of differentially expressed-aberrantly methylated hub genes in breast cancer. J Cancer. 2019;10(26):6618–6634. doi:10.7150/jca.33433
19. Chen J, Wang Z, Shen X, et al. Identification of novel biomarkers and small molecule drugs in human colorectal cancer by microarray and bioinformatics analysis. Mol Genet Genomic Med. 2019;7(7). doi:10.1002/mgg3.713.
20. Jin G, Wang H, Hu Z, et al. Potentially functional polymorphisms of EXO1 and risk of lung cancer in a Chinese population: a case-control analysis. Lung Cancer. 2008;60(3):340–346. doi:10.1016/j.lungcan.2007.11.003
21. Hsu N-Y, Wang H-C, Wang C-H, et al. Lung cancer susceptibility and genetic polymorphisms of Exo1 gene in Taiwan. Anticancer Res. 2009;29(2):725–730.
22. Matakidou A, El Galta R, Webb EL, et al. Genetic variation in the DNA repair genes is predictive of outcome in lung cancer. Hum Mol Genet. 2007;16(19):2333–2340. doi:10.1093/hmg/ddm190
23. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi:10.1158/2159-8290.CD-12-0095
24. Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi:10.1016/S1476-5586(04)80047-2
25. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
26. Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–w102. doi:10.1093/nar/gkx247
27. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e10. doi:10.1158/0008-5472.CAN-17-0307
28. Szklarczyk D, Gable A, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D13. doi:10.1093/nar/gky1131
29. Stelzer G, Rosen N, Plaschkes I, et al. The geneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54(1):
30. Azim HA, Peccatori FA, Brohée S, et al. RANK-ligand (RANKL) expression in young breast cancer patients and during pregnancy. Breast Cancer Res. 2015;17(1):24. doi:10.1186/s13058-015-0538-7
31. Meador CB, Hata AN. Acquired resistance to targeted therapies in NSCLC: updates and evolving insights. Pharmacol Ther. 2020;210.
32. de Sousa JF, Serafim RB, de Freitas LM, et al. DNA repair genes in astrocytoma tumorigenesis, progression and therapy resistance. Genet Mol Biol. 2020;43.
33. Sun J, Wang C, Zhang Y, et al. Genomic signatures reveal DNA damage response deficiency in colorectal cancer brain metastases. Nat Commun. 2019;10(1):3190. doi:10.1038/s41467-019-10987-3
34. Tomimatsu N, Mukherjee B, Harris JL, et al. DNA-damage-induced degradation of EXO1 exonuclease limits DNA end resection to ensure accurate DNA repair. J Biol Chem. 2017;292(26):10779–10790. doi:10.1074/jbc.M116.772475
35. Zhang M, Zhao D, Yan C, et al. Associations between nine polymorphisms in EXO1 and cancer susceptibility: a systematic review and meta-analysis of 39 case-control studies. Sci Rep. 2016;6.
36. Li Z, Sang M, Tian Z, et al. Identification of key biomarkers and potential molecular mechanisms in lung cancer by bioinformatics analysis. Oncol Lett. 2019;18(5):4429–4440. doi:10.3892/ol.2019.10796
37. Gao Y, Hayes RB, Huang W-Y, et al. DNA repair gene polymorphisms and tobacco smoking in the risk for colorectal adenomas. Carcinogenesis. 2011;32(6):882–887. doi:10.1093/carcin/bgr071
38. de Sousa JF, Torrieri R, Serafim RB, et al. Expression signatures of DNA repair genes correlate with survival prognosis of astrocytoma patients. Tumour Biol. 2017;39(4):1010428317694552. doi:10.1177/1010428317694552
39. Liberti S, Andersen S, Wang J, et al. Bi-directional routing of DNA mismatch repair protein human exonuclease 1 to replication foci and DNA double strand breaks. DNA Repair. 2011;10(1):73–86. doi:10.1016/j.dnarep.2010.09.023
40. Chen X, Paudyal S, Chin R, et al. PCNA promotes processive DNA end resection by Exo1. Nucleic Acids Res. 2013;41(20):9325–9338. doi:10.1093/nar/gkt672
41. Viktorsson K, De Petris L, Lewensohn RJB, et al. The role of p53 in treatment responses of lung cancer. Biochem Biophys Res Commun. 2005;331(3):868–880. doi:10.1016/j.bbrc.2005.03.192
42. Liu H, Liu Y, Yang F, et al. Acetylation of MORC2 by NAT10 regulates cell-cycle checkpoint control and resistance to DNA-damaging chemotherapy and radiotherapy in breast cancer. Nucleic Acids Res. 2020;48(7):3638–3656. doi:10.1093/nar/gkaa130
43. He D, Li T, Sheng M, et al. Exonuclease 1 (Exo1) participates in mammalian non-homologous end joining and contributes to drug resistance in ovarian cancer. Med Sci Monit. 2020;26.
44. Li R, Gu J, Heymach JV, et al. Hypoxia pathway genetic variants predict survival of non-small-cell lung cancer patients receiving platinum-based chemotherapy. Carcinogenesis. 2017;38(4):419–424. doi:10.1093/carcin/bgx014
45. Sansregret L, Vanhaesebroeck B, Swanton C. Determinants and clinical implications of chromosomal instability in cancer. Nat Rev Clin Oncol. 2018;15(3):139–150. doi:10.1038/nrclinonc.2017.198
46. Liu D, Keijzers G, Rasmussen LJ. DNA mismatch repair and its many roles in eukaryotic cells. Mutat Res Rev Mutat Res. 2017;773:174–187.
47. Varella-Garcia M. Chromosomal and genomic changes in lung cancer. Cell Adh Migr. 2010;4(1):100–106. doi:10.4161/cam.4.1.10884
48. Fu F, Zhang Y, Gao Z, et al. Development and validation of a five-gene model to predict postoperative brain metastasis in operable lung adenocarcinoma. Int J Cancer. 2020;147(2):584–592. doi:10.1002/ijc.32981
49. Amin NS, Nguyen MN, Oh S, et al. Exo1-dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol Cell Biol. 2001;21(15):5142–5155. doi:10.1128/MCB.21.15.5142-5155.2001
50. Bolderson E, Tomimatsu N, Richard DJ, et al. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 2010;38(6):1821–1831. doi:10.1093/nar/gkp1164
51. Shenker RF, McTyre ER, Ruiz J, et al. The effects of smoking status and smoking history on patients with brain metastases from lung cancer. Cancer Med. 2017;6(5):944–952. doi:10.1002/cam4.1058
52. Wu SY, Xing F, Sharma S, et al. Nicotine promotes brain metastasis by polarizing microglia and suppressing innate immune function. J Exp Med. 2020;217(8):217. doi:10.1084/jem.20191131
53. Tan Q, Wang G, Huang J, et al. Epigenomic analysis of lung adenocarcinoma reveals novel DNA methylation patterns associated with smoking. Onco Targets Ther. 2013;6:1471–1479. doi:10.2147/OTT.S51041
54. Zhou L, Gao HF, Liu DS, et al. Gene expression profiling of brain metastatic cell from triple negative breast cancer: understanding the molecular events. Gene. 2018;640:21–27. doi:10.1016/j.gene.2017.10.019
55. Zeni E, Mazzetti L, Miotto D, et al. Macrophage expression of interleukin-10 is a prognostic factor in nonsmall cell lung cancer. Eur Respir J. 2007;30(4):627–632. doi:10.1183/09031936.00129306
56. Villanueva MT. Cancer immunotherapy: macrophages steal the show. Nat Rev Drug Discov. 2017;16(7):455. doi:10.1038/nrd.2017.126
57. Donnem T, Hald SM, Paulsen EE, et al. Stromal CD8+T-cell density—A promising supplement to tnm staging in non–small cell lung cancer. Clin Cancer Res. 2015;21(11):2635–2643. doi:10.1158/1078-0432.CCR-14-1905
58. Zhang Y, Gallastegui N, Rosenblatt JD. Regulatory B cells in anti-tumor immunity. Int Immunol. 2015;27(10):521–530. doi:10.1093/intimm/dxv034
59. Saha T, Sundaravinayagam D, Di Virgilio M. Charting a DNA repair roadmap for immunoglobulin class switch recombination. Trends Biochem Sci. 2020. doi:10.1016/j.tibs.2020.10.005
60. Chi X, Li Y, Qiu X. V(D)J recombination, somatic hypermutation and class switch recombination of immunoglobulins: mechanism and regulation. Immunology. 2020;160(3):233–247. doi:10.1111/imm.13176
61. Bouaziz JD, Yanaba K, Venturi GM, et al. Therapeutic B cell depletion impairs adaptive and autoreactive CD4+ T cell activation in mice. Proc Natl Acad Sci U S A. 2007;104(52):20878–20883. doi:10.1073/pnas.0709205105
62. Zhu W, Germain C, Liu Z, et al. A high density of tertiary lymphoid structure B cells in lung tumors is associated with increased CD4+T cell receptor repertoire clonality. Oncoimmunology. 2015;4(12):e1051922. doi:10.1080/2162402X.2015.1051922
63. Xu F, Chen JX, Yang XB, et al. Analysis of lung adenocarcinoma subtypes based on immune signatures identifies clinical implications for cancer therapy. Mol Ther Oncolytics. 2020;17:241–249. doi:10.1016/j.omto.2020.03.021
64. Rosenthal R, Cadieux EL, Salgado R, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature. 2019;567(7749):479–485. doi:10.1038/s41586-019-1032-7
65. Germain C, Gnjatic S, Tamzalit F, et al. Presence of B cells in tertiary lymphoid structures is associated with a protective immunity in patients with lung cancer. Am J Respir Crit Care Med. 2014;189(7):832–844. doi:10.1164/rccm.201309-1611OC
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.