Back to Journals » Infection and Drug Resistance » Volume 11
Evaluation of the GenoType MTBDRplus and MTBDRsl for the detection of drug-resistant Mycobacterium tuberculosis on isolates from Beijing, China
Authors Jian J , Yang X, Yang J, Chen L
Received 7 June 2018
Accepted for publication 18 July 2018
Published 1 October 2018 Volume 2018:11 Pages 1627—1634
DOI https://doi.org/10.2147/IDR.S176609
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Jiyong Jian,1–3 Xinyu Yang,4 Jun Yang,5 Liang Chen1–3
1Clinical Laboratory Medicine, Beijing Shijitan Hospital, Capital Medical University, Beijing, China; 2Peking University Ninth School of Clinical Medicine, Beijing, China; 3Beijing Key Laboratory of Urinary Cellular Molecular Diagnostics, Beijing, China; 4Central Laboratory, Beijing Research Institute for Tuberculosis Control, Beijing, China; 5Department of Sterilized supplying, PLA 306 Hospital, Beijing, China
Background: The incidence of tuberculosis (TB), especially multidrug-resistant TB (MDR-TB) and extensively drug-resistant TB (XDR-TB), continues to increase alarmingly worldwide. Molecular line probe assays (LPAs) are endorsed by the World Health Organization for the fast detection of MDR-TB and XDR-TB. The aim of this study was to evaluate the performance of LPAs in China.
Methods: We analyzed MDR-TB and XDR-TB in 96 isolates from Beijing by using culture-based drug susceptibility testing (DST) and LPAs to compare the detection rate of the two methods.
Results: Compared to phenotypic DST, the GenoType® MTBDRplus and MTBDRsl, respectively, showed a sensitivity of 98.7% and a specificity of 88.9% for detection of rifampicin resistance, 82.1% and 94.4% for isoniazid, 89.7% and 94.4% for levofloxacin, 60.0% and 98.7% for amikacin/capreomycin, and 57.5% and 98.2% for ethambutol. The sensitivity and specificity of LPAs, respectively, were 80.8% and 100% for MDR-TB and 50.0% and 97.6% for XDR-TB. Mutations in codon S531L of the rpoB gene and S315T1 of the KatG gene were dominated in MDR-TB strains. The most frequently observed mutations were in codon A90V of the gyrA gene, A1401G of the rrs gene, and M306V of the embB gene, according to the MTBDRsl results.
Conclusion: Our study showed that, in combination with phenotypic DST, application of the LPAs might be an efficient and reliable supplementary DST assay for rapid susceptibility screening of MDR-TB and XDR-TB. Using LPAs in countries with high MDR/XDR burden allows for appropriate and timely treatment, which will reduce transmission rates and morbidity, and improve treatment outcomes in patients.
Keywords: Mycobacterium tuberculosis, MDR-TB, XDR-TB, line probe assay, phenotypic drug susceptibility testing
Introduction
Tuberculosis (TB) remains a global public health threat, especially in developing countries. According to the global TB report of the World Health Organization (WHO), in 2016, TB was responsible for the deaths of 1.67 million people. And it generated 10.4 million new cases in this year, of which 490,000 were diagnosed as multidrug-resistant (MDR-TB) and 8,014 were diagnosed as extensively drug-resistant (XDR-TB).1 In China, a country with high TB burden, ~110,000 new MDR-TB and 8,200 new XDR-TB cases emerge annually according to a national survey held in 2007.2 According to another report, about 55% of MDR-TB cases in China remain unidentified.3 MDR-TB is resistant to both the two most powerful anti-TB drugs, namely rifampicin (RFP) and isoniazid (INH), and requires treatment with a second-line regimen. XDR-TB is defined as MDR-TB plus resistance to at least one fluoroquinolone (FQ) and a second-line injectable drug (SLID), including amikacin (AM), capreomycin (CPM), and kanamycin (KM), the two most important classes of medicines in an MDR-TB regimen.
It is critical to use susceptibility testing to screen for resistance to specific antibiotics for the detection of MDR-TB and XDR-TB so that treatment regimen could be designed specifically on the basis of the detection results. Conventional culture-based phenotypic drug susceptibility testing (DST) is considered to be the gold standard for determining drug resistance. It is important for the confirmation of MDR-TB and the assessment of drug resistance to second-line and new drugs in the management of MDR-TB and XDR-TB. However, it takes months for phenotypic DST to yield final results.4 Molecular-based assays designed to detect specific drug resistance-encoding mutations in Mycobacterium tuberculosis (MTB) have the advantage of achieving results within 48 hours, much faster than conventional DST. In 2008, WHO endorsed the molecular test GenoType® MTBDRplus (Hain Lifescience, Nehren, Germany) for rapid detection of resistance to RFP and INH. The assay detects mutations in the rpoB gene for RFP resistance, in the katG gene for high-level INH resistance, and in the inhA regulatory region gene for low-level INH resistance.5 In May 2016, WHO recommended using GenoType MTBDRsl (Hain Lifescience) to detect mutations in the gyrA, rrs, and embB genes for the screening of resistance to FQ, SLID, and ethambutol (EMB) in the diagnosis of XDR-TB among MDR-TB patients.6
In order to rapidly detect MDR-TB and XDR-TB, WHO endorsed line probe assays (LPAs) of MTBDRplus and MTBDRsl for the detection of RFP, INH, FQ, SLID, and EMB resistance in acid-fast bacilli smear-positive sputum or MTB cultures in 2017. The aim of this study was to compare the diagnostic performance of the MTBDRplus and MTBDRsl assays with the gold standard phenotypic DST in the detection of MDR-TB and XDR-TB, among culture isolates obtained from patients in Beijing.
Materials and methods
Study design
The evaluation of the GenoType MTBDRplus v1.0 and MTBDRsl v1.0 assays were conducted at the Beijing Shijitan Hospital, Beijing Research Institute for Tuberculosis Control, and PLA 306 Hospital. The study was approved by the ethics committees of all the above participants. A total of 96 MTB isolates were collected from Beijing Research Institute for Tuberculosis Control between 2015 and 2016, including 78 MDR-TB (including 12 XDR-TB) and 18 randomly chosen fully susceptible isolates based on phenotypic DST. The MTB isolates in our study were part of the routine hospital laboratory procedure. We compared the performance of the MTBDRplus v1.0 and MTBDRsl v1.0 assays with that of phenotypic MTB DST in susceptibility testing of first- and second-line anti-TB drugs.
DST
Phenotypic DST was performed against RFP, INH, levofloxacin (LFX), AM, CPM, and EMB using the standard version of the WHO proportion method on Lowenstein–Jensen medium (L-J)7,8 and considered the gold standard for the detection of resistance. The following critical concentrations of drugs recommended by WHO for testing of drug-resistant TB using proportion method DST were used: RFP 40 µg/mL, INH 0.2 µg/mL, LFX 2 µg/mL, AM 30 µg/mL, CPM 40 µg/mL, and EMB 2 µg/mL. Susceptibility is determined by comparison of growth on the control medium with growth on a drug-containing medium.
LPAs
The MTBDRplus and the MTBDRsl assays were performed directly on the MTB isolates according to the manufacturer’s instructions. The person performing the tests was blinded to the proportion method DST. It was considered as a valid result if all six expected control bands appeared correctly; otherwise, the result was considered invalid. The absence of at least one of the wild-type bands or the presence of bands indicating a mutation in each drug resistance-related gene implied that the sample was resistant to the specific antibiotic. When all the wild-type probes of a gene stained positive and there were no detectable mutations within the region examined, the sample was considered susceptible to the respective antibiotic.
Quality control
The fully susceptible MTB H37Rv reference strain was used as quality control (QC) for proportion method DST and LPA. This QC strain is susceptible to first- and second-line drugs tested in this study.
Statistical analysis
Sensitivity, specificity, negative predictive value (NPV), positive predictive value (PPV), and agreement of LPA compared to proportion method DST were calculated. The precision of the estimates was reported using 95% CIs. P≤0.05 was considered statistically significant. Agreement between the two methods was assessed using the kappa statistic. All data were analyzed by SPSS version 19.0 (SPSS Inc, Chicago, IL, USA).
Results
Phenotypic DST results
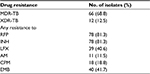
Table 1 summarizes drug susceptibility patterns of isolates included in the study. The isolate collection (n=96) contained 78 MDR-TB (81.3%), of which 12 isolates were XDR-TB (12.5%). Among these isolates, 78 (81.3%) were resistant to RFP, 78 (81.3%) were resistant to INH, 39 (40.6%) were resistant to LFX, 11 (11.5%) were resistant to AM, 18 (18.8%) were resistant to CM, and 40 (41.7%) were resistant to EMB by conventional phenotypic DST.
Performance of GenoType MTBDRplus and MTBDRsl assay
The performance of GenoType MTBDRplus has been summarized in Table 2. The sensitivity for RFP resistance was determined as 98.7% (95% CI 96.2–100), specificity 88.9% (95% CI 72.8–100), PPV 97.5% (95% CI 93.9–100), NPV 94.1% (95% CI 81.6–100), and diagnostic efficacy 96.9%; for INH resistance, sensitivity 82.1% (95% CI 73.3–90.8), specificity 94.4% (95% CI 82.7–100), PPV 98.5% (95% CI 95.4–100), NPV 54.8% (95% CI 36.3–73.4), and diagnostic efficacy 84.4%; and for MDR-TB, sensitivity 80.8% (95% CI 71.8–89.7), specificity 100% (95% CI 100–100), PPV 100% (95% CI 100–100), NPV 54.5% (95% CI 36.6–72.5), and diagnostic efficacy 84.4%, compared to phenotypic DST.
The performance of GenoType MTBDRsl is also shown in Table 2. The sensitivity for detecting LFX resistance was 89.7% (95% CI 79.8–99.7), specificity 96.5% (95% CI 91.6–100), PPV 94.6% (95% CI 87.0–100), NPV 93.2% (95% CI 86.6–99.8), and diagnostic efficacy 93.8%; for AM/CPM resistance, sensitivity 60.0% (95% CI 36.5–83.5), specificity 98.7% (95% CI 96.1–100), PPV 92.3% (95% CI 75.5–100), NPV 90.4% (95% CI 83.9–96.8), and diagnostic efficacy 90.6%; for EMB resistance, sensitivity 57.5% (95% CI 41.5–73.5), specificity 98.2% (95% CI 94.6–100), PPV 95.8% (95% CI 87.2–100), NPV 76.4% (95% CI 66.3–86.4), and diagnostic efficacy 81.2%; and for XDR-TB, sensitivity 50.0% (95% CI 16.8–83.2), specificity 97.6% (95% CI 94.3–100), PPV 75.0% (95% CI 36.3–100), NPV 93.2% (95% CI 87.8–98.6), and diagnostic efficacy 91.7%.
Detection of mutations associated with drug resistance using MTBDRplus and MTBDRsl assay
The distribution of mutations is summarized in Table 3. Mutations in rpoB conferring resistance to RFP were detected in 82.3% (79/96) of the isolates. The RFP-resistant isolates displayed different mutations: 58.2% (46/79) of the isolates had mutation at position S531L, 10.1% (8/79) of the isolates had mutation at position D516V, 8.9% (7/79) of the isolates had mutation at position H526Y, while 15 isolates had mutations only at the wild-type probes. Among the 15 isolates with mutations detected only at the wild-type probes, seven isolates had mutation at rpoB WT7, five isolates at WT8, two isolates at WT3 and WT4, and one isolate at WT2 and WT3. According to the kit manufacturer’s recommendation, these isolates were considered resistant and indicated the presence of a less common or rare mutation.
A total of 67.7% (65/96) of the isolates showed mutations in katG gene or inhA promoter region indicating that they were resistant to INH. There were four isolates that showed mutations at both katG and inhA gene. Among the 65 INH-resistant strains, 73.8% (48/65) had mutation in the katG gene with amino acid change of S315T1, indicating high-level resistance, while 24.6% (16/65) of the strains had mutation in the inhA gene, C15T, indicating low-level resistance. One isolate was detected with mutation at wild-type probes.
There were 38.5% (37/96) isolates resistant to LFX as tested by LPA. The majority of the gyrA mutations, 37.8% (14/37), were observed at D94G. Other gyrA mutations detected by the assay were at A90V (13/37; 35.1%), at D94A (8/37; 21.6%), and at S91P (5/37; 13.5%). A total of 59.5% (22/37) mutations in gyrA were detected at codon 94.
The isolates had been detected with 12 defined mutations and one undefined mutations in the rrs gene among the 13/96 (13.5%) isolates by LPA. The most frequently observed mutation (12/13; 92.3%) for AM/CPM resistance was rrs MUT1 (A1401G).
EMB resistance was detected in 24 (25.0%) of 96 isolates, in which the mutation M306V was the most prevalent (15/24; 62.5%), followed by the mutation M306I (9/24; 37.5%) in embB gene.
Discussion
Drug-resistant TB poses a great threat to TB control programs worldwide. Early diagnosis and effective treatment require a sensitive and specific diagnostic tool. According to the WHO, LPA is an optimal susceptibility testing of MTB in the selection of anti-TB drugs for an effective treatment regimen.1 In this study, we evaluated the diagnostic accuracy of MTBDRplus and MTBDRsl using culture isolates in Beijing.
MTBDRplus showed high sensitivity and specificity for the detection of susceptibility to RFP (sensitivity 98.7% and specificity 88.9%). The sensitivity for susceptibility to RFP was in concordance with the high sensitivity of the MTBDRplus assay in the range of 95%–100%.9–11 In this study, the S531L mutation in rpoB was the most frequent (58.2%), followed by the D516V mutation (10.1%). This is similar to the frequencies reported in other studies.11,12 The high detection rate can be explained by the fact that the mutations responsible for RFP resistance are mainly located in the 81-bp hot-spot region and mutations outside this location are rare and are associated with low-level resistance.13,14
The sensitivity and specificity of MTBDRplus for the detection of susceptibility to INH were 82.1% and 94.4%, respectively. In our study, the sensitivity for INH resistance was much lower than 86%–100%15,16 and higher than 69.9%, as shown in another study using clinical isolates in Eastern China.17 In total, 65 isolates were detected with INH resistance by the GenoType MTBDRplus, among which 73.8% carried a mutation at the S315T1 codon of the katG gene and 24.6% with the mutation C15T in the inhA regulatory region. As previously described by several authors, the most common mutation involved in INH resistance is the S315T substitution in katG, which is related to high-level INH resistance.18,19 The most prevalent mutation in the inhA gene detected using LPA in our study was C15T with loss of WT1, which confers low-level INH resistance; this is also supported by previous studies.20,21 The relatively low sensitivity to detect INH resistance for MTBDRplus is due to the complex molecular basis, which involves mutations in more than one gene or gene complex, such as the katG, inhA, and kasA genes and the intergenic region of the oxyR-ahpC complex.22,23
Previous studies have shown that the sensitivity of GenoType MTBDRsl assay ranged from 75.6% to 90.6% for detecting FQ resistance.24–26 In this study, the sensitivity of the MTBDRsl assay for detecting LFX resistance in clinical strains was 89.7%. FQ resistance in MTB is ascribed mainly to gyrA mutations, with 57.5% of mutations detected at codon 94% and 31.5% at codon 90.27 Consistent with published data, the highest frequency of mutations conferring FQ resistance was observed in gyrA codon 94 (59.5%), followed by codon 90 (13/37, 35.1%). Our study showed that the most prevalent mutation pattern in gyrA was the D94G mutation, followed by the A90V and D94A mutation, which is in agreement with other studies.24,28 Given that mutation probes for gyrB are not included in the assay, it is possible that some isolates may have had mutations in these positions as well but could not be detected.
Our study showed a low sensitivity of the LPA for the detection of resistance to SLID (60.0%), which was much lower than other reports (86.7%, 100%)29,30 and was similar to Zeng’s report.28 The variable results may be ascribed to the fact that geographically different MTB lineages can result in different gene mutation patterns. Interestingly, cross-resistance to AM, KM, and CPM had been reported.31 The predominant rrs gene mutation was A1401G (92.3%) detected by MTBDRsl assay according to our study. Consistent with published data,30,32 rrs MUT1 A1401G was the most frequently observed mutation among tested isolates. In this study, we found eight isolates which MTBDRsl assay showed sensitive to SLID, while the phenotypic DST indicated that they were resistant to CPM but sensitive to AM. The rrs1401 mutation alone was not found with sufficient frequency to be detected in more than 70%–80% of global MTB strains resistant to AM and CPM, while the eis promoter, tlyA and gidB appeared to be involved in the resistance to AM and CPM.33
The sensitivity of the MTBDRsl in the detection of EMB resistance was 57%–69.2%.24–26 Our results confirmed the poor performance of embB mutations in the detection of EMB resistance. A recent meta-analysis showed a similar sensitivity with the MTBDRsl assay for detecting EMB resistance (55%).34 The most common mutations detected in embB by the MTBDRsl assay were M306V in 62.5% and M306I in 37.5% of EMB-resistant isolates, which corresponded to previously published report.35 This suggests that the significance of mutations in this codon is limited and it is necessary to identify other mutations conferring resistance to this drug. Huang et al identified several mutations in embB other than that at codon 306.35 At the same time, recent results on the proficiency testing of DST in supranational TB reference laboratories highlighted a lack of consistency in DST results for EMB, and the reproducibility was also found to be poor.36
As a rapid diagnosis of MDR-TB or even XDR-TB is of high importance for patient outcome and of high epidemiological importance, we evaluated detection performance of the GenoType MTBDRplus and MTBDRsl assays on the MTB isolates. It takes less than one day for the assays to get information about the MTB resistance pattern, but takes two weeks for the conventional DST testing. The sensitivity (80.8%) for the detection of MDR-TB in the present study was much lower than previous report.10 We also found that GenoType MTBDRsl was specific (97.6%) for the diagnosis of XDR-TB, although the sensitivity was very low (50.0%), as reported in previous studies.17
There are several limitations to our work. MTBDRplus v1.0 and MTBDRsl v1.0 were used, which had recently been succeeded by a new iteration (v2.0).37,38 The new MTBDRplus v2.0 test shows a higher analytical sensitivity when compared with the original MTBDRplus, which allows this new version to be performed on both smear-positive and smear-negative clinical specimens. GenoType MTBDRsl v2.0 was redesigned based on GenoType MTBDRsl v1.0 and accommodated additional mutations for the molecular detection of resistance to FQ involving gyrA and gyrB and SLID resistance covering both rrs and eis genes.39 A further limitation was our sample size, particularly the second-line anti-TB drugs-resistant group was small. Finally, both LPA assays were only tested on the MTB isolates, but not patient’s samples. It takes up to seven weeks for positive MTB cultures, while rapid and safe diagnosis of MDR-TB and XDR-TB is essential for the adequate treatment of patients.
In conclusion, discordance still exists between conventional and LPA approaches in resistance testing. Further investigations are required in the negative results as resistance to second-line drugs may still be present but undetected by LPA assays. Even though MTBDRsl had suboptimal diagnostic sensitivity for FQ and SLID, MTBDRsl remained to play an important supplementary role for the rapid detection of FQ and SLID resistance, given that phenotypic DST has a prolonged within-laboratory turn-around-time and is technically challenging. At the same time, given the poor performance of the MTBDRsl assay for detecting EMB resistance, this assay can be used neither for detecting nor for ruling out EMB resistance accurately, and clinicians should use sequencing-based methods for identifying other mutations conferring resistance to this drug or await the results of phenotypic DST before deciding on changes in treatment regimens. We suggest that LPAs can be used as a supplementary method for the detection of MDR-TB and XDR-TB in high-burden countries to improve treatment outcomes in patients. Reducing the time to diagnosis, commencing appropriate therapy timeously, and preventing transmission of drug-resistant strains are major advantages of LPAs.
Acknowledgments
We thank all of the persons involved in this study for their essential work and support. We also wish to thank bioMérieux Shanghai Co. Limited for their technical support for this study.
This study was funded by Beijing Municipal Administration of Hospitals’ Ascent Plan (grant no. DFL20150701).
Disclosure
The authors report no conflicts of interest in this work.
References
World Health Organization. Global Tuberculosis Report. 2017. Organization World Health; 2017. | ||
Zhao Y, Xu S, Wang L, et al. National survey of drug-resistant tuberculosis in China. N Engl J Med. 2012;366(23):2161–2170. | ||
World Health Organization. Global Tuberculosis Report. 2014. World Health Organization; 2014. | ||
Bwanga F, Hoffner S, Haile M, Joloba ML. Direct susceptibility testing for multi drug resistant tuberculosis: a meta-analysis. BMC Infect Dis. 2009;9:67. | ||
Piersimoni C, Olivieri A, Benacchio L, Scarparo C. Current perspectives on drug susceptibility testing of Mycobacterium tuberculosis complex: the automated nonradiometric systems. J Clin Microbiol. 2006;44(1):20–28. | ||
Theron G, Peter J, Richardson M. The diagnostic accuracy of the Genotype® MTBDRsI assay for the detection of resistance to second-line anti-tuberculosis drugs. Cochrane Database Syst Rev. 2014;10:CD010705. | ||
Canetti G, Fox W, Khomenko A, et al. Advances in techniques of testing mycobacterial drug sensitivity, and the use of sensitivity tests in tuberculosis control programmes. Bull World Health Organ. 1969;41(1):21–43. | ||
Canetti G, Froman S, Grosset J, et al. Mycobacteria: laboratory methods for testing drug sensitivity and resistance. Bull World Health Organ. 1963;29:565–578. | ||
Asante-Poku A, Otchere ID, Danso E, et al. Evaluation of GenoType MTBDRplus for the rapid detection of drug-resistant tuberculosis in Ghana. Int J Tuberc Lung Dis. 2015;19(8):954–959. | ||
Yadav RN, Singh BK, Sharma SK, et al. Comparative evaluation of GenoType MTBDRplus line probe assay with solid culture method in early diagnosis of multidrug resistant tuberculosis (MDR-TB) at a tertiary care centre in India. PLoS One. 2013;8(9):e72036. | ||
Maningi NE, Malinga LA, Antiabong JF, Lekalakala RM, Mbelle NM3. Comparison of line probe assay to BACTEC MGIT 960 system for susceptibility testing of first and second-line anti-tuberculosis drugs in a referral laboratory in South Africa. BMC Infect Dis. 2017;17(1):795. | ||
Bang D, Andersen SR, Vasiliauskienė E, Rasmussen EM. Performance of the GenoType MTBDRplus assay (v2.0) and a new extended GenoType MTBDRsl assay (v2.0) for the molecular detection of multi- and extensively drug-resistant Mycobacterium tuberculosis on isolates primarily from Lithuania. Diagn Microbiol Infect Dis. 2016;86(4):377–381. | ||
Heep M, Brandstätter B, Rieger U, et al. Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J Clin Microbiol. 2001;39(1):107–110. | ||
Hillemann D, Weizenegger M, Kubica T, Richter E, Niemann S. Use of the genotype MTBDR assay for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2005;43(8):3699–3703. | ||
Hillemann D, Rüsch-Gerdes S, Richter E. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol. 2007;45(8):2635–2640. | ||
Felkel M, Exner R, Schleucher R, et al. Evaluation of Mycobacterium tuberculosis drug susceptibility in clinical specimens from Nigeria using genotype MTBDRplus and MTBDRsl assays. Eur J Microbiol Immunol. 2013;3(4):252–257. | ||
Liu Q, Li GL, Chen C, et al. Diagnostic Performance of the GenoType MTBDRplus and MTBDRsl Assays to Identify Tuberculosis Drug Resistance in Eastern China. Chin Med J. 2017;130(13):1521–1528. | ||
Ramaswamy SV, Reich R, Dou SJ, et al. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2003;47(4):1241–1250. | ||
van Soolingen D, de Haas PE, van Doorn HR, et al. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in the Netherlands. J Infect Dis. 2000;182(6):1788–1790. | ||
Zhang Z, Lu J, Liu M, et al. Genotyping and molecular characteristics of multidrug-resistant Mycobacterium tuberculosis isolates from China. J Infect. 2015;70(4):335–345. | ||
Tolani MP, D’Souza DT, Mistry NF. Drug resistance mutations and heteroresistance detected using the GenoType MTBDRplus assay and their implication for treatment outcomes in patients from Mumbai, India. BMC Infect Dis. 2012;12:9. | ||
Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79(1):3–29. | ||
Somoskovi A, Parsons LM, Salfinger M. The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respir Res. 2001;2(3):164–168. | ||
Brossier F, Veziris N, Aubry A, Jarlier V, Sougakoff W. Detection by GenoType MTBDRsl test of complex mechanisms of resistance to second-line drugs and ethambutol in multidrug-resistant Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2010;48(5):1683–1689. | ||
Hillemann D, Rüsch-Gerdes S, Richter E. Feasibility of the GenoType MTBDRsl assay for fluoroquinolone, amikacin-capreomycin, and ethambutol resistance testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol. 2009;47(6):1767–1772. | ||
Kiet VS, Lan NT, An DD, Dd A, et al. Evaluation of the MTBDRsl test for detection of second-line-drug resistance in Mycobacterium tuberculosis. J Clin Microbiol. 2010;48(8):2934–2939. | ||
Li J, Gao X, Luo T, et al. Association of gyrA/B mutations and resistance levels to fluoroquinolones in clinical isolates of Mycobacterium tuberculosis. Emerg Microbes Infect. 2014;3(3):e19. | ||
Zeng X, Jing W, Zhang Y, et al. Performance of the MTBDRsl Line probe assay for rapid detection of resistance to second-line anti-tuberculosis drugs and ethambutol in China. Diagn Microbiol Infect Dis. 2017;89(2):112–117. | ||
Lee YS, Lee BY, Jo KW, Shim TS. Performance of the GenoType MTBDRsl assay for the detection second-line anti-tuberculosis drug resistance. J Infect Chemother. 2017;23(12):820–825. | ||
Ajbani K, Nikam C, Kazi M, et al. Evaluation of genotype MTBDRsl assay to detect drug resistance associated with fluoroquinolones, aminoglycosides and ethambutol on clinical sediments. PLoS One. 2012;7(11):e49433. | ||
Maus CE, Plikaytis BB, Shinnick TM. Molecular analysis of cross-resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2005;49(8):3192–3197. | ||
Via LE, Cho SN, Hwang S, et al. Polymorphisms associated with resistance and cross-resistance to aminoglycosides and capreomycin in Mycobacterium tuberculosis isolates from South Korean Patients with drug-resistant tuberculosis. J Clin Microbiol. 2010;48(2):402–411. | ||
Georghiou SB, Magana M, Garfein RS, et al. Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: a systematic review. PLoS One. 2012;7(3):e33275. | ||
Cheng S, Cui Z, Li Y, Hu Z. Diagnostic accuracy of a molecular drug susceptibility testing method for the antituberculosis drug ethambutol: a systematic review and meta-analysis. J Clin Microbiol. 2014;52(8):2913–2924. | ||
Huang WL, Chi TL, Wu MH, Jou R. Performance assessment of the GenoType MTBDRsl test and DNA sequencing for detection of second-line and ethambutol drug resistance among patients infected with multidrug-resistant Mycobacterium tuberculosis. J Clin Microbiol. 2011;49(7):2502–2508. | ||
van Deun A, Wright A, Zignol M, Weyer K, Rieder HL. Drug susceptibility testing proficiency in the network of supranational tuberculosis reference laboratories. Int J Tuberc Lung Dis. 2011;15(1):116–124. | ||
Crudu V, Stratan E, Romancenco E, et al. First evaluation of an improved assay for molecular genetic detection of tuberculosis as well as rifampin and isoniazid resistances. J Clin Microbiol. 2012;50(4):1264–1269. | ||
Tagliani E, Cabibbe AM, Miotto P, et al. Diagnostic Performance of the New Version (v2.0) of GenoType MTBDRsl Assay for Detection of Resistance to Fluoroquinolones and Second-Line Injectable Drugs: a Multicenter Study. J Clin Microbiol. 2015;53(9):2961–2969. | ||
HAIN LifeScience. GenoType MTBDRsl VER 2.0 instructions for use. Document IFU-317A-01. Nehren, Germany: HAIN LifeScience; 2015. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.