Back to Journals » Infection and Drug Resistance » Volume 16
Evaluation of the Diagnostic Performance of mNGS in Detecting Intra-Abdominal Infections of the Emergency Department Patients
Authors Zheng L, Kang Z, Wang R, Lv M, Gao Z , Xu H, Wang M
Received 8 November 2022
Accepted for publication 10 February 2023
Published 13 March 2023 Volume 2023:16 Pages 1421—1432
DOI https://doi.org/10.2147/IDR.S396699
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Liang Zheng,1,* Zhoujun Kang,1,* Ru Wang,2 Meng Lv,2 Zhirui Gao,1 Haizhou Xu,1 Meitang Wang1
1Emergency Department, Changhai Hospital Affiliated to Navy Medical University, Shanghai, People’s Republic of China; 2Genoxor Medical Science and Technology Inc., Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Haizhou Xu; Meitang Wang, Emergency Department, Changhai Hospital Affiliated to Navy Medical University, Changhai Road No. 168, Yangpu District, Shanghai, 200433, People’s Republic of China, Tel +86-21-81873891, Email [email protected]; [email protected]
Purpose: Intra-abdominal infections (IAI) are gradually becoming common in the emergency department, though the incidence is low and the prognosis is fair, as the symptoms are similar to other intra-abdominal diseases, rapid and accurate diagnosis of the causative agents is essential for clinical management. This study aimed to evaluate the diagnostic performance of metagenomic next-generation sequencing (mNGS) in detecting IAI in the emergency department.
Patients and Methods: This was a retrospective, single-centered study including patients admitted to the emergency department from January 1st, 2021 to August 31st, 2022 with diagnosis of IAI. The comparison between mNGS and microbial culture using paracentesis fluid samples was performed to evaluate the diagnostic performance of mNGS for IAI. Meanwhile, paracentesis fluid and peripheral blood mNGS were compared to explore the sample specificity. Further, the microbial community structure of the patients with pyogenic liver abscesses (PLA) was analyzed.
Results: Thirty-four IAI patients including 23 with pyogenic liver abscesses (PLA), 3 with parapancreatic abscesses, and 8 with other IAI were included in this study. Compared with the conventional microbial culture of paracentesis fluid, mNGS using paracentesis fluid detected more positive cases of IAI (93.75% vs 81.25%), and identified more species of pathogens, especially in obligate anaerobes and viral pathogens. Peripheral blood mNGS presented a relatively high consistency with the paracentesis fluid mNGS (91% mutual positive). The microbial community structure of PLA patients with diabetes is less diverse than that of those without diabetes. Patients with diabetes are at high risk of PLA caused by Klebsiella pneumonia.
Conclusion: mNGS has advantages in detecting IAI in the emergency department, and peripheral blood mNGS can be a non-invasive choice for early diagnosis.
Keywords: metagenomic next-generation sequencing, mNGS, abdominal infection, pyogenic liver abscesses, PLA, emergency department
Introduction
An intra-abdominal abscess is a collection of pus or infected fluid located inside or near the liver, kidneys, pancreas, spleen, or other abdominal organs.1 Unlike skin abscesses with obvious signs of redness and swelling,2 intra-abdominal abscesses occur less frequently and are often difficult to identify, of which patients may have symptoms including but not limited to pain, fever, chills, and loss of appetite.3,4 Bacterial infections are the main cause of intra-abdominal abscesses,5,6 and viruses, fungi, and parasites may cause abscesses in few cases.7,8 Clinical management of intra-abdominal abscesses necessitates surgical drainage and antibiotic treatment. Currently, intra-abdominal abscesses like pyogenic liver abscesses (PLA) are gradually common in the emergency department. Studies revealed that the incidence of PLA varies from 3.6 to 17.1 per 100,000 individuals among regions, with the main causative pathogens of Streptococcus species, Staphylococcus species, Escherichia coli, and Klebsiella pneumonia.9–13 Therefore, accurate and timely diagnosis of the causative agents is essential for targeted antibiotic therapy.
The culture of aspirated pus is the gold standard test for the diagnosis of abscesses, and blood culture can also help determine the etiology.14 However, the positive rate of microbial culture is low in clinical, and it usually takes several days or more to isolate the species.15 With the advancement of high throughput sequencing technologies and their application in abdominal abscesses diagnosis, these culture-independent technologies showed superior diagnostic performance. A study used nanopore sequencing to analyze the crude liver abscess aspirates, which achieved real-time pathogen identification.16 Metagenomic next-generation sequencing (mNGS) has also been applied to detect pathogenic microorganisms of PLA in the laboratory and in clinical settings.17,18 Compared to traditional microbial culture, mNGS possesses advantages in a broad spectrum of pathogen detection, shortening turnaround time, and metagenomic analyses of antibiotic resistance prediction could be employed for better treatment and prognosis.19,20 Although cases of mNGS applied in the emergency department17,21 were reported, cohort studies are needed to explore its diagnostic performance.
In this study, we assessed the diagnostic performance of mNGS in detecting intra-abdominal abscesses in the emergency department, and performed metagenomics analyses of pathogenic microorganisms detected through the mNGS method, aiming to provide referential experiences for clinical practice.
Materials and Methods
Ethical Statement
This study was conducted in accordance with the Declaration of Helsinki and its revisions, and approved by the ethical committee of Changhai Hospital. Written informed consent was obtained from all patients who participated in this study or from the parent or legal guardian of the patient under 18 years of age.
Patients
In this study, we retrospectively reviewed a cohort of IAI patients in the emergency department of our hospital from January 1st, 2021 to August 31st, 2022. Patients of suspected IAI which routinely sent for mNGS assay in the emergency department were included in this study. Patients of other infection were excluded. All patients confirmed the diagnosis of the abscess through imaging examinations (CT and ultrasound) and laboratory tests (microbial culture and mNGS) combined with clinical manifestations. Figure 1 shows the design of this study.
 |
Figure 1 Flow chart of the study design. |
Microbial Culture
According to the culture procedures for bacteria and fungi in the diagnostic laboratory of Changhai Hospital, the sample culture was performed using routine isolation media, including blood agar, eosin methylene-blue (EMB) agar, chocolate agar, Mueller-Hinton agar, sabouraud dextrose agar, and cooked meat medium (Oxoid, UK). Plates were incubated at 37°C in 5–10% CO2 for 24–48 hours using BC60 automated culture system (Autobio Diagnostics Co., Ltd., China). The strains were then isolated and identified the species using the VITEK-2 Compact Instrument (bioMérieux, France).
mNGS Sequencing and Analysis
Samples collected were sent to Genoxor Medical Science and Technology Inc. (Shanghai, China) for mNGS sequencing. In the laboratory, after pre-processing, TIANamp Micro DNA Kit (DP316, Tiangen Biotech, Beijing, China) was applied to extract the DNA of the samples following the manufacturer’s instructions. Then, DNA fragmentation, end-repair, adapter ligation, and PCR amplification were performed by step to construct the DNA libraries. The quality of the libraries was assessed by Agilent 2100. The libraries were sequenced on the NextSeqTM 550DX sequencer according to the manufacturer’s instructions.
Raw data were subjected to a quality control process to remove low-quality sequences by Trimmomatic. Human genome sequences were excluded by Bowtie2. An in-house Perl script was used to remove identical duplicate reads considered as technical artifacts by PCR. Retained sequences were processed using Kraken for taxonomic classification and a custom k-mer database was constructed using 51,543 genomes of ~27,000 species from the NCBI assembly databases.
Alpha diversity measured by the Shannon diversity index was used to assess the complexity of taxonomic diversity. Beta diversity measured using Principal Coordinate Analysis (PCoA) based on the Bray–Curtis distance was used to assess the compositional similarity. Receiver operating characteristic (ROC) curves were calculated and analyzed. Linear discriminant-analysis effect-size (LEfSe) analysis was performed to determine the significantly different taxa between groups.
Statistical Analysis
Statistical analysis was conducted by GraphPad Prism Version 9 (GraphPad Software, USA). Mean ± standard deviation (SD) was used for describing continuous variables. A χ2 or Fisher’s exact test and a t-test were applied for analyses of categorical variables and continuous variables, respectively. A P-value <0.05 was considered statistically significant.
Results
General Characteristics and Clinical Features of the Patients
From January 1st, 2021 to August 31st, 2022, a total of 34 patients admitted to the emergency department of our hospital were diagnosed with IAI through imaging and were included in this research, as shown in Table 1, when admitted, 23 patients (67.6%) presented fevers to different extents, 4 of which with a body temperature over 39°C. In addition to fever, clinical symptoms like abdominal pain, chills, and diarrhea were also recorded. Results of laboratory examinations showed elevated levels of white blood cell (WBC), neutrophil (NE), C-reactive protein (CRP), and procalcitonin (PCT) among the patients, indicating potential infections. The general characteristics of the patients and laboratory examination results are listed in Table S1.
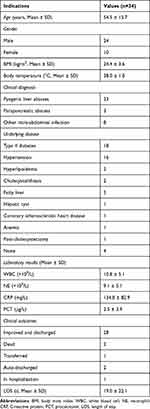 |
Table 1 General Characteristics of the Patients |
Microbial Recovered from the Patients
Clinical samples of paracentesis fluid and peripheral blood were collected and sent for microbial culture and mNGS, respectively, to detect the causative pathogens. As shown in Figure 2, thirteen species of pathogens were identified through microbial culture, with 6 species of G+ bacteria, and 7 species of G- bacteria. The paracentesis fluid mNGS detected 15 species of pathogens, with 4 species of G+ bacteria, 11 species of G- bacteria, and 3 species of obligate anaerobes. Eighteen species of pathogens were detected through the peripheral blood mNGS, with 4 species of G+ bacteria, 13 species of G- bacteria, 3 species of obligate anaerobes, and 2 viruses. Compared with the conventional microbial culture, more kinds of pathogens were detected through mNGS. Meanwhile, bacteria such as Bacteroides fragilis, Parabacteroides distasonis, Bacteroides thetaiotaomicron, etc, difficult to culture had also been identified via the mNGS method. In addition to bacterial pathogens, viral pathogens such as Human betaherpesvirus 5 and Human gammaherpesvirus 4 were also detected through the peripheral blood mNGS.
 |
Figure 2 Pathogens identified through microbial culture (A), paracentesis fluid mNGS (B) and peripheral blood mNGS (C). |
Comparison of Pathogen Detection Performance Between mNGS and Microbial Culture
Further, to assess the diagnostic performance of mNGS, we compared the results of thirty-two patients who underwent the paracentesis fluid culture and paracentesis fluid mNGS simultaneously. Twenty-six positive cases were detected through the microbial culture, with a positive rate of 81.25%, and 30 (93.75%) were detected positive in paracentesis fluid mNGS. According to our results, more positive cases were detected by mNGS than by culture, while the difference between culture and mNGS was not significant. Among all these positive cases, 5 patients were detected with polymicrobial infection through culture, and 8 were through paracentesis fluid mNGS (Figure 3A). The turnaround time of mNGS (1 day) was greatly shorter than that of microbial culture (5.03 ± 0.63 days, Mean ± SD). As shown in Figure 3B, we compared the consistency of the two methods. Regarding the overall results, mNGS and culture were both negative in 3% of cases; 16% of cases were detected positive only through mNGS, and 3% only through culture. For mutual positive cases, 56% of which were detected with completely consistent pathogens. Taking the culture results as the standard, the sensitivity and specificity of paracentesis fluid mNGS were 92.51%, and 16.67%, respectively.
Comparison of Pathogen Detection Performance Between Paracentesis Fluid mNGS and Peripheral Blood mNGS
Meanwhile, the peripheral blood mNGS was also performed to explore whether a difference exists between samples. Thirty (93.75%) of thirty-two samples were detected positive in the peripheral blood mNGS, and 9 cases were detected with polymicrobial infections (Figure 4A). A slight superiority of the peripheral blood mNGS was noted compared to the paracentesis fluid mNGS in detecting polymicrobial infection cases, while no significant difference was noted. As shown in Figure 4B, a relatively high concordance between these two samples was presented. 3% of cases were detected negative in both the paracentesis fluid mNGS and the peripheral blood mNGS; 3% of cases were detected positive only through the peripheral blood samples, and 3% were only through the paracentesis fluid samples. For mutual positive cases, 67% of which were detected with completely consistent pathogens.
 |
Figure 4 Diagnostic performance of mNGS between different samples. (A) Comparison of positive rate of the two samples. (B) The consistency between the two samples. |
mNGS-Guided Antimicrobial Therapy and Clinical Outcomes
All patients were prescribed antibiotics for anti-infective treatment, with empirical medications like meropenem, tigecycline, piperacillin sodium/tazobactam sodium, etc. After mNGS detection, the therapeutic regimens for 20 patients had been adjusted for better treatment. For Patient 6, meropenem+tigecycline has been altered to metronidazole because of the detected Streptococcus anginosus through mNGS. For Patient 18, imipenem/cilastatin was applied for the empirical anti-infective treatment initially; however, Acinetobacter baumannii complex was identified through mNGS as well as the anti-microbial resistance genes were detected, and the medication was changed to meropenem+tigecycline. After treatment, laboratory examinations showed that the WBC, NE, CRP, and PCT levels of the patients decreased (Figure 5). Twenty-eight patients were improved and discharged, two were dead, two were discharged automatically and two are still in hospitalization (Table 1).
 |
Figure 5 Comparisons of laboratory examinations between before surgery and before discharge. (A) WBC. (B) NE. (C) CRP. (D) PCT. ***Denotes P-value <0.0001. |
Community Structure of the Peripheral Blood and Paracentesis Fluid Microbiota of Pyogenic Liver Abscesses
In this study, PLA (23 of 34) was the most common disease of the abdominal abscess patients in the emergency department, and 17 of the PLA patients also had the underlying condition of diabetes. After certain treatment, blood sugar levels decreased from 11.53 ± 4.10 mmol/L to 9.57 ± 3.71 mmol/L (Mean ± SD). With diabetes being an important risk factor for PLA, we compared and analyzed the detected metagenomic data from peripheral blood and paracentesis fluid samples of the patients, to explore the differences in the microbial communities between PLA patients with and without diabetes.
In the peripheral blood samples, the top 10 abundant species in each group are shown in Figure 6A. The Shannon index (Figure 6B) showed that the richness and diversity of the microbiota within the diabetes group is lower than that of no_diabetes group, and beta diversity (Figure 6C) in the diabetes group is also lower than that from no_diabetes group (P < 0.001). Linear discriminant-analysis effect-size (LEfSe) analyses (Figure 6D) found four species of bacteria significantly different between the diabetes group and no_diabetes group. However, as Figure 6E shows, the microbial community of the peripheral blood samples from the patients with diabetes was not significantly different from that in the no_diabetes group. The ROC curves in Figure 6F indicates the difference of Klebsiella pneumonia between the two groups, with an AUC of 0.79.
Meanwhile, we compared the microbial community in the paracentesis fluid samples between PLA patients with and without diabetes. Figure 7A shows the top 10 abundant species in each group, with Klebsiella pneumonia being the major bacterium in patients with diabetes. The Shannon index of the diabetes group is lower than no_diabetes group (Figure 7B), indicating the richness and diversity within the diabetes group are less than the other group. Beta diversity (Figure 7C) in the diabetes group is also lower than that from no_diabetes group (P < 0.005). Two species of bacteria were found significantly different between diabetes group and no_diabetes group (Figure 7D). From Figure 7E, the microbial community of the paracentesis fluid samples from the patients with diabetes was different from that in the no_diabetes group. The ROC curves in Figure 7F indicates the difference in Klebsiella pneumonia between the two groups, with an AUC of 0.86. In our study, the detection rate of Klebsiella pneumonia in the diabetes group was 93.33%, and 50% in the no_diabetes group. This difference was further confirmed by ROC curves.
Discussion
In this study, we assessed the diagnostic performance of mNGS in detecting intra-abdominal abscesses of patients in the emergency department. Based on the current study, the mNGS assay exhibited high consistency with the microbial culture method in detecting intra-abdominal infections. Compared with routine microbial culture, the mNGS assay could identify more positive cases and species of causative agents, provide with metagenomic analyses for guiding clinical management, and greatly shorten the turnaround time (1 day vs 5.03 ± 0.63 days, Mean ± SD). Meanwhile, comparison between mNGS of different samples revealed that the peripheral blood mNGS can help early diagnosis and treatment. Further, the analysis of microbial community of PLA help improve our knowledge of this disease.
As reported, mNGS exhibited superior performance in detecting various infections: in detecting central nervous system infection, mNGS assay offered faster turnaround time compared to conventional testing, and could identify pathogens that cannot be identified from conventional testing;22 in detecting infections of respiratory system,23 urinary system,24 etc., mNGS method also presented similar advantages over conventional culture. Although the superiority in our cohort is not significant, which could be reasonable as many causative pathogens in our study can be easily incubated and isolated through microbial culture, such as Klebsiella pneumonia. Furthermore, more species have been identified through mNGS method, like Bacteroides fragilis and Parabacteroides distasonis, and several viral pathogens, owing to the advantage to sequence all the genomes in the micro-environment unbiasedly.15 Although viral pathogens detected in this study had scarcely effect in infection management of the patients due to clinical examinations, they should be taken into consideration with prudence if relevant symptoms presented. Another strength of applying mNGS is to guide the anti-infective treatment, as it could provide with metagenomic analyses like antibiotic resistance prediction.25 Culture-based methods can provide insights into anti-infective therapy guides. However, these methods are often limited to microorganisms easily to culture. The initial anti-infective therapeutics for 20 patients in this study have been altered after mNGS test, especially in patients for which mNGS results presented strains with antimicrobial resistance genes.
In addition, we performed mNGS simultaneously in peripheral blood samples and paracentesis fluid samples to evaluate the difference in diagnostic performance between different samples. As shown in our study, the peripheral blood mNGS results were highly consistent with that of paracentesis fluid mNGS. 91% of the cases were detected positive in both samples, with 67% of which identified the same pathogens and 23% identifying the partial same pathogens. A probable explanation is that pathogens spread into the blood with the infection processing. Blood samples have been widely applied to infection diagnosis, as the advantages of accessibility and less-invasive. A study has emphasized the utility of blood mNGS in bloodstream infection detection in patients with severe pneumonia.26 Another research27 revealed that the peripheral blood mNGS as a rapid and non-invasive method for detecting Aspergillus. While a study from Catherine A Hogan et al to evaluate the clinical impact of plasma cell-free DNA mNGS for the diagnosis of infectious diseases found that currently the impact is limited and remains further studies.28 From the results of our study, for patients with abdominal abscesses, although aspiration of paracentesis fluids for laboratory and clinical examinations is essential, the peripheral blood mNGS can be a favorable choice to assist in earlier detection and diagnosis. In research and practice, microbial contamination from exogenous and endogenous sources remained a challenge to the reliability of mNGS results. Moreover, colonizers detected by mNGS assays may also influence the clinical interpretation of mNGS results, highlighting the requirement for the interpretation of mNGS reports carefully, and the combination of relevant laboratory and clinical tests. Compared with conventional culture, mNGS exerted superior sensitivity in detecting pathogens, and with a correspondingly higher false-positive rate. In this study, pathogens detected in both peripheral blood mNGS and paracentesis fluids mNGS help to exclude contaminants and may be more instructive for clinical management.
Further, as PLA is one of the severe abscesses, and gradually become common in the emergency department,29,30 metagenomic analyses of the microbial community were performed both in the paracentesis fluid and peripheral blood samples, aiming to improve the understanding of this disease. The richness and diversity of the microbial community in patients with diabetes are lower than that without diabetes, indicating the effects of underlying conditions on microbiota diversity. In PLA patients complicated with diabetes, Klebsiella pneumonia is the most dominant pathogen. While in PLA patients with other underlying diseases, the community structure is more diverse and complicated. Based on our results, patients with diabetes are at high risk of PLA caused by Klebsiella pneumonia. Many studies have revealed that PLA became common in patients with diabetes. Research from Du et al31 and Li et al32 showed the proportion of PLA patients complicated with diabetes was 26.87% and 36.6%, respectively, and comorbid diabetes had effects on the complications and short-term mortality of the PLA patients. A meta-analysis from Chan et al33 revealed that Klebsiella pneumonia PLA has lower mortality than non-Klebsiella pneumonia PLA. Meanwhile, extrahepatic syndrome caused by Klebsiella pneumonia may affect the lungs, eyes, and central nervous system,34 developing metastatic complications including meningitis, endophthalmitis, etc. Under these conditions, rapid detection of causative pathogens is essential for earlier diagnosis and treatment, so as to improve clinical outcomes.
Nonetheless, our study has several limitations. This is a single-center retrospective study with a relatively small sample size. Hence, the prospective study with a large sample size is required for further exploration of the diagnostic value of mNGS applied to intra-abdominal infections in the emergency department.
Conclusion
To conclude, the mNGS method exhibited high consistency with the microbial culture in detecting IAI and offered advantages in detecting viral pathogens. Considering the emergency department’s high demand for time efficiency, applying mNGS assays could greatly shorten the turnaround time. Moreover, peripheral blood mNGS can be a noninvasive supplement for earlier detection and diagnosis.
Data Sharing Statement
The metagenomic sequencing data comprising microbial reads have been deposited at the NCBI SRA database, under BioProject accession PRJNA898073.
Ethics Approval and Informed Consent
This study was conducted in accordance with the Declaration of Helsinki and its revisions, and approved by the ethical committee of Changhai hospital. Written informed consent was obtained from all patients who participated in this study or from the parent or legal guardian of the patient under 18 years of age.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the National Natural Science Foundation of China (Grant 81570073).
Disclosure
Ru Wang and Meng Lv are affiliated with Genoxor Medical Science and Technology Inc. The authors declare that they have no other conflicts of interest in this work.
References
1. Mehta NY, Copelin IE. Abdominal abscess. In: StatPearls. Treasure Island (FL): StatPearls Publishing LLC; 2022.
2. Schmitz GR, Gottlieb M. Managing a cutaneous abscess in the emergency department. Ann Emerg Med. 2021;78(1):44–48. doi:10.1016/j.annemergmed.2020.12.003
3. Longworth S, Han J. Pyogenic liver abscess. Clin Liver Dis. 2015;6(2):51–54. doi:10.1002/cld.487
4. Tung CC, Chen FC, Lo CJ. Splenic abscess: an easily overlooked disease? Am Surg. 2006;72(4):322–325. doi:10.1177/000313480607200409
5. Marra A, Hillejan L, Ukena D. Therapie von Lungenabszessen [Management of Lung Abscess]. Zentralbl Chir. 2015;140(Suppl 1):S47–53. German. doi:10.1055/s-0035-1557883
6. Takayanagi N, Kagiyama N, Ishiguro T, Tokunaga D, Sugita Y. Etiology and outcome of community-acquired lung abscess. Respiration. 2010;80(2):98–105. doi:10.1159/000312404
7. Schneider M, Kobayashi K, Uldry E, Demartines N, Golshayan D, Halkic N. Rhizomucor hepatosplenic abscesses in a patient with renal and pancreatic transplantation. Ann R Coll Surg Engl. 2021;103(4):e131–e135. doi:10.1308/rcsann.2020.7125
8. Lee JY, Park JW, Park JY. Liver abscess caused by cytomegalovirus in a patient with acquired immunodeficiency syndrome. Infect Chemother. 2020;54(4):803–807.
9. Meddings L, Myers RP, Hubbard J, et al. A population-based study of pyogenic liver abscesses in the United States: incidence. Mortality Temporal Trends. 2010;105(1):117–124.
10. Zimmermann L, Wendt S, Lübbert C, Karlas T. Epidemiology of pyogenic liver abscesses in Germany: analysis of incidence, risk factors and mortality rate based on routine data from statutory health insurance. United European Gastroenterol J. 2021;9(9):1039–1047. doi:10.1002/ueg2.12132
11. Jepsen P, Vilstrup H, Schønheyder HC, Sørensen HT. A nationwide study of the incidence and 30-day mortality rate of pyogenic liver abscess in Denmark, 1977–2002. Aliment Pharmacol Ther. 2005;21(10):1185–1188. doi:10.1111/j.1365-2036.2005.02487.x
12. Cerwenka H. Pyogenic liver abscess: differences in etiology and treatment in Southeast Asia and Central Europe. World J Gastroenterol. 2010;16(20):2458–2462. doi:10.3748/wjg.v16.i20.2458
13. Tsai F-C, Huang Y-T, Chang L-Y, Wang J-T. Pyogenic liver abscess as endemic disease, Taiwan. Emerging Infectious Diseases. 2008;14(10):1592. doi:10.3201/eid1410.071254
14. Miller JM, Binnicker MJ, Campbell S, et al. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2018 update by the infectious diseases society of America and the American society for microbiology. Clin Infect Dis. 2018;67(6):e1–e94. doi:10.1093/cid/ciy381
15. Han D, Li Z, Li R, Tan P, Zhang R, Li J. mNGS in clinical microbiology laboratories: on the road to maturity. Crit Rev Microbiol. 2019;45(5–6):668–685. doi:10.1080/1040841X.2019.1681933
16. Gong L, Huang YT, Wong CH, et al. Culture-independent analysis of liver abscess using nanopore sequencing. PLoS One. 2018;13(1):e0190853. doi:10.1371/journal.pone.0190853
17. Xie J, Zhu Z. A case report of pyogenic liver abscess caused by hypervirulent Klebsiella pneumoniae diagnosed by metagenomic next-generation sequencing. J Int Med Res. 2021;49(7):3000605211032793. doi:10.1177/03000605211032793
18. Zeng S, Yan WQ, Wu XM, Zhang HN. Case report: diagnosis of Klebsiella pneumoniae invasive liver abscess syndrome with purulent meningitis in a patient from pathogen to lesions. Front Med. 2021;8:714916. doi:10.3389/fmed.2021.714916
19. Gu W, Miller S, Chiu CY. Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol. 2019;14(1):319–338. doi:10.1146/annurev-pathmechdis-012418-012751
20. Wilson MR, Sample HA, Zorn KC, et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med. 2019;380(24):2327–2340. doi:10.1056/NEJMoa1803396
21. Wu J, Wu Y, Huang M. Metagenomic next-generation sequencing helped diagnose scrub typhus without eschar: a case report. Int J Infect Dis. 2020;90:1–4. doi:10.1016/j.ijid.2019.10.020
22. Kawada J, Okuno Y, Torii Y, et al. Identification of viruses in cases of pediatric acute encephalitis and encephalopathy using next-generation sequencing. Sci Rep. 2016;6:33452. doi:10.1038/srep33452
23. Li H, Gao H, Meng H, et al. Detection of pulmonary infectious pathogens from lung biopsy tissues by metagenomic next-generation sequencing. Front Cell Infect Microbiol. 2018;8. doi:10.3389/fcimb.2018.00205
24. Zhou Y, Wylie KM, Feghaly REE, et al. Metagenomic approach for identification of the pathogens associated with diarrhea in stool specimens. J Clin Microbiol. 2016;54(2):368–375. doi:10.1128/JCM.01965-15
25. Sukhum KV, Diorio-Toth L, Dantas G. Genomic and metagenomic approaches for predictive surveillance of emerging pathogens and antibiotic resistance. Clin Pharmacol Ther. 2019;106(3):512–524. doi:10.1002/cpt.1535
26. Chen J, Zhao Y, Shang Y, et al. The clinical significance of simultaneous detection of pathogens from bronchoalveolar lavage fluid and blood samples by metagenomic next-generation sequencing in patients with severe pneumonia. J Med Microbiol. 2021;70:1. doi:10.1099/jmm.0.001259
27. Ma X, Zhang S, Xing H, et al. Invasive pulmonary aspergillosis diagnosis via peripheral blood metagenomic next-generation sequencing. Front Med. 2022;9:751617. doi:10.3389/fmed.2022.751617
28. Hogan CA, Yang S, Garner OB, et al. Clinical impact of metagenomic next-generation sequencing of plasma cell-free DNA for the diagnosis of infectious diseases: a multicenter retrospective cohort study. Clin Infect Dis. 2020;72(2):239–245. doi:10.1093/cid/ciaa035
29. Liu L, Chen W, Lu X, Zhang K, Zhu C. Pyogenic liver abscess: a retrospective study of 105 cases in an emergency department from East China. J Emerg Med. 2017;52(4):409–416. doi:10.1016/j.jemermed.2016.09.026
30. Yin D, Ji C, Zhang S, et al. Clinical characteristics and management of 1572 patients with pyogenic liver abscess: a 12-year retrospective study. Liver International. 2021;41(4):810–818. doi:10.1111/liv.14760
31. Du Z, Zhou X, Zhao J, et al. Effect of diabetes mellitus on short-term prognosis of 227 pyogenic liver abscess patients after hospitalization. BMC Infect Dis. 2020;20(1):145. doi:10.1186/s12879-020-4855-9
32. Li W, Chen H, Wu S, Peng J. A comparison of pyogenic liver abscess in patients with or without diabetes: a retrospective study of 246 cases. BMC Gastroenterol. 2018;18(1):144. doi:10.1186/s12876-018-0875-y
33. Chan KS, Chia CTW, Shelat VG. Demographics, radiological findings, and clinical outcomes of Klebsiella pneumonia vs. Non-Klebsiella pneumoniae pyogenic liver abscess: a systematic review and meta-analysis with trial sequential analysis. Pathogens. 2022;11(9):976. doi:10.3390/pathogens11090976
34. Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. 2012;12(11):881–887. doi:10.1016/S1473-3099(12)70205-0
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



