Back to Journals » OncoTargets and Therapy » Volume 10
Evaluation of p53 gene expression and prognosis characteristics in uveal melanoma cases
Received 11 March 2017
Accepted for publication 22 April 2017
Published 12 July 2017 Volume 2017:10 Pages 3429—3434
DOI https://doi.org/10.2147/OTT.S136785
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Carlos E Vigil
Haojie Liu, Ming Zhou
Department of Ophthalmology, Affiliated Zhongshan Hospital of Dalian University, Dalian, China
Objective: The objective of this study was to evaluate the correlation between the expression of p53 gene and the prognosis after local excision in uveal melanoma.
Materials and methods: Real-time polymerase chain reaction (RT-PCR) test and Western blot were used to detect the expression of p53 in C918, MUM-2B, and D78 cell lines at the levels of messenger RNA (mRNA) and protein. Immunohistochemistry staining was done in the tissues of 68 patients, which were diagnosed with uveal melanoma. Furthermore, the effects of p53 protein on the invasion abilities of both the cell lines were studied by transinfection of p53 small interfering RNA. The clinical and prognostic data regarding the effect of p53 protein on the patient’s prognosis were calculated and further analyzed by Kaplan–Meier univariate analysis method.
Results: The results of RT-PCR and Western blot revealed that p53 mRNAs were highly expressed in C918 and MUM-2B cells. The high expression rate of p53 among the 88 uveal melanoma tissues was 77.27%. Transinfection of p53 serine could inhibit the expression of p53 in uveal melanoma and the invasion ability of the cells. This study found that the high expression of p53 and the prognosis of uveal melanoma patients were statistically correlated.
Conclusion: The expression of p53 protein in uveal melanoma was unusual and was associated with the invasion ability of uveal melanoma. This indicates that the highest expression of p53 protein indicates worse prognosis of uveal melanoma patients.
Keywords: uveal melanoma, PCR, prognosis, Western blot, immunohistochemistry
Introduction
Melanoma of the uveal tract is the most common primary intraocular tumor in adults. Despite advances in the treatment of primary tumor, there are currently no successful treatment options for metastatic tumor. Uveal melanoma has the high ability of invasion and metastasis.1,2 The most common location of initial metastatic site is liver, followed by lung and soft tissues.3 The prognosis for metastatic uveal melanoma is poor with a 1-year overall mortality of 80%–87%, further increasing to 92% at 2 years.4
p53 gene is considered to be the most frequently and commonly altered gene, which shows mutations in various tumors. Therefore, considering p53 as the bookmaker is of important clinical significance of the tumors.5 Earlier, various authors and researchers first identified the p53 gene, as a kind of cancer protein antigen, then as a cancer gene, and finally as a tumor suppressor gene. Further elaborative research and clinical trials concluded that the mutation of p53 gene could result in tumorigenesis or cell transformation. The mutant protein of p53 can be considered as a uveal melanoma promoter. In addition, the wild-type p53 gene is a kind of tumor suppressor gene; hence, mutation of p53 gene will induce tumorigenesis.6
Various authors concluded that the expression of p53 protein in breast cancer and oral squamous cell carcinoma is closely correlated with the prognosis of patients.7 Usually, tumors of highly malignant potential and poor prognosis show higher expression of p53 protein. Therefore, the expression of p53 protein can be considered as the biomarker of the prognosis of the breast cancer and the oral squamous cell carcinoma.8
The aim of this research was to evaluate the effects of the expression of p53 protein in uveal melanoma on the prognosis after the local excision. Studies have found that p53 mutant protein is highly expressed in the cell lines and tissues of uveal melanoma. Therefore, gene silencing technique could be used to inhibit the expression of p53 protein and to identify the effect of p53 protein on the invasion ability of uveal melanoma. In addition, the study aimed to evaluate the effect of the expression of p53 protein on operation prognosis.
Materials and methods
All the research materials, including the laboratory parameters used in the study, were the uveal melanoma cells (C918 and MUM-2B) and normal human uveal melanoma cell (D78; Chinese Academy of Sciences Cell Bank, Shanghai, China); Dulbecco’s Modified Eagle’s Medium (DMEM) culture (Invitrogen, Waltham, MA, USA); fetal bovine serum (FBS; HyClone Laboratories, Logan, UT, USA); Lipofectamine 2000, primer, and miR-655 mimics (Invitrogen, Carlsbad, CA, USA); real-time polymerase chain reaction kits (Ambion, Austin, TX, USA); p53, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primary antibody, horseradish peroxidase (HRP)-conjugated secondary antibody 1QAZ (Proteintech Group, Inc, Wuhan, China); transwell chambers (Corning Inc., Corning, NY, USA); immunohistochemical staining kit SP-9001 (Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China); and primers and small interfering RNAs (siRNAs; Shanghai GenePharma, Shanghai, China). Before starting the study, an ethical approval was obtained from the Dalian Ethics committees and Institutional Review Board, and written informed patient consent regarding the study was also collected from all the subjects. The study was registered under a trial number of DU/CTN/2014/87965.
Eighty-eight local excised tissues of uveal melanoma patients and 20 local tissues of the corresponding normal sides of the cancer were also included in the clinical research. The samples of uveal melanoma tissues were confirmed as uveal melanoma by histopathological examination.
Cell culture
C918, MUM-2B, and D78 cells were cultured by DMEM culture (including 10% FBS) at 37°C in the culture box with 5% CO2. The culture medium was changed after 48 hours. The subculture was digested when the degree of fusion had reached 80%.
RNA extraction and detection of the expression of p53 messenger RNA (mRNA) in cells
One microgram of total RNA was collected from every group to conduct reverse transcription according to the instructions given by the manufacturer on the kits. p53 primer was added and the expression of p53 mRNA was detected by quantitative polymerase chain reaction (PCR). GAPDH was selected as internal parameter. p53 primer and GAPDH primer sequences are shown in Table 1. The PCR conditions were as follows: initial denaturation at 94°C for 5 minutes, followed by 30 cycles for amplification (94°C for 30 seconds, 56°C for 30 seconds, and 72°C for 1.5 minutes) and 72°C for 5 minutes (Figure 1).
  | Table 1 RT-PCR primer sequence and silence sequence |
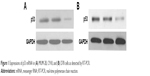  | Figure 1 Expressions of p53 mRNA in (A) MUM-2B, C918, and (B) D78 cells as detected by RT-PCR. |
Detection of the expression of p53 protein in cells by Western blot
MUM-2B, C918, and D78 cells were cultured separately with cell lysis buffer. Total protein was extracted as per the instructions mentioned on the kits. Bicinchoninic acid was used to detect the protein concentration. Proteins were separated via 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene difluoride membrane. After antibody incubation steps, a band intensity was detected via chemiluminescence, imaged, and quantified. After sealing with 5% bovine serum albumin, p53 and GAPDH antibody were diluted to 5 μg/mL and 1/10,000, respectively, and added into the cell culture medium, which was incubated for overnight at 4°C, and then for 2 hours by HRP as the secondary antibody. Enhanced chemiluminescence was used to develop images in darkroom. Afterward, the images were scanned and the records were maintained by taking the photographs (Figure 2).
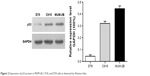  | Figure 2 Expression of p53 protein in MUM-2B, C918, and D78 cells as detected by Western blot. |
Detection of the expression of p53 protein in pathological tissues by immunohistochemistry
All tissue samples were routinely fixed in 10% neutral formalin and embedded in paraffin. Sections of 4 μm were cut from one representative block of each case and collected onto glass slides. The slides were deparaffined, rehydrated in graded alcohols, and submitted to heat-induced antigen retrieval with citrate 10 mM, pH 6.0 in a pressure cooker (Pascal; Dako, Carpentaria, CA, USA) for p53. Endogenous peroxidase activity block was performed with 8% hydrogen peroxide in methanol.
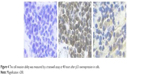
Primary antibodies were diluted in phosphate-buffered saline and incubated for 18 hours at 4°C (anti-p53). Primary antibodies used for immunohistochemical evaluation included an antihuman polyclonal p53 antibody (1:500, clone CM1; Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.). The secondary antibody complex used was the EnVision kit (Dako). Diaminobenzidine was used as a chromogen. Slides were counterstained with Mayer’s hematoxylin, washed in running water for 5 minutes, dehydrated, and mounted in a synthetic medium. Then the positive controls and negative controls were standardized (Figure 3). According to the following grading standard, positive cell staining number was classified (no staining or <5% as 0 point, <40% staining as 1 point, >40% as 2 points, >60% as 3 points) into two groups: low p53 expression group (≤1 point) and high p53 expression group (≥2 points). Grading results were statistically analyzed.
  | Figure 3 Expression of p53 protein in pathological tissues as detected by immunohistochemistry method (400×). |
Cell-transinfected p53 siRNA
Transinfection experiment included three groups: control group, negative control (NC) siRNA group, and p53 siRNA group. For MUM-2B, C918, and D78 cells at the logarithmic growth phase, pancreation was used to digest the cells and prepare unicellular suspension. Six-well culture plates were used to incubate the cells. After adherence, 2 μg siRNA was transinfected in every well. Transinfection was carried out according to the instructions mentioned on the Lipofectamine 2000 kit. For transfection of one well in the six-well plate, 500 μL Opti-MEM I reduced serum medium was prepared containing 2.5 μg of enhanced GFP mRNA and the respective amount of Lipofectamine 2000. The components were gently mixed by pipetting. The transfection mixture was then incubated at room temperature for 20 minutes to generate lipoplexes for transfection. Cells were washed with 500 μL Dulbecco’s phosphate-buffered saline per well, and the transfection mixture was pipetted into the well. After 4 hours of incubation at 37°C and 5% CO2, the transfection mixture was replaced by 1 mL complete cell culture medium. Cells were cultivated for 24 hours in the cell incubator and analyzed using flow cytometry.
Effect of p53 siRNA on p53 protein expression and the invasion ability of cells
After 48 hours of transinfection, the expressions of p53 protein in cells of three groups were detected as described earlier.
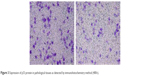
In this section, the transwell chambers were included, and the data were analyzed in all the three groups: control group, NC siRNA group, and p53 siRNA group. Operations and preparations were conducted according to the given instructions. About 100 μL of unicellular suspension with the concentration of 4×105/mL and 100 μL of serum-free culture medium were evenly added to the upper compartment, whereas 500 μL of culture medium with 30% FBS was added to the lower compartment. Nonmigrated cells were removed from the upper side of the filters using cotton swabs, whereas migrated cells in the lower face of the filters were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. For invasion assay, transwell inserts were coated with metrigel. Images of five random ×10 fields were captured from each well and counted under a light microscope (Figure 4).
  | Figure 4 The cell invasion ability was measured by a transwell assay at 48 hours after p53 overexpression in cells. |
Correlation between the expression of p53 in uveal melanoma and the prognosis
All patients were divided into two groups: the low p53 expression group and the high p53 expression group, which were further correlated. Both the groups had matched age, gender, smoking history, family history, and other factors. The effect of p53 protein expression on the prognosis of uveal melanoma patients was analyzed according to the follow-up results.
Statistical analysis
The collected research data were expressed as mean ± standard deviation. SPSS version 17.0 software (SPSS Inc., Chicago, IL, USA) was used for single-factor analysis of variance. Clinical prognosis data were analyzed by Kaplan–Meier survival analysis method. P-value <0.05 was considered as statistically significant.
Results
Expression of p53 mRNA in D78, C918, and MUM-2B cells
p53 mRNA showed higher expression in C918 and MUM-2B cells. Comparison of the expression of p53 mRNA among D78 control, C918, and MUM-2B groups revealed statistically significant differences with a P-value of <0.01.
Expression of p53 protein in MUM-2B, C918, and D78 cells
Western blot detection results showed that the expression rate of p53 protein in C918 and MUM-2B cells was relatively high compared with D78 control group. Comparison of the expression of p53 protein among D78 control, C918, and MUM-2B groups revealed significant differences with a P-value of <0.01.
Expression of p53 protein in the tissues of uveal melanoma
Immunohistochemisty results revealed that p53 was a nuclear protein. Therefore, the positivist was shown as yellow cell-stained nucleus. Among the tissues of 88 patients, 62 patients showed a positive expression of p53 (77.27%) (Table 2).
  | Table 2 Expression of p53 protein in normal tissues and uveal melanoma tissues |
Effect of p53 siRNA on the invasion ability of cells
The results revealed that after transinfection of C918 cells by siRNA, the number of cells that passed through between the NC siRNA group and the control group revealed no statistical significance. However, when the number of cells that passed through between the p53 siRNA group and the control group was compared, the results revealed a statistically significant P-value of <0.01. MUM-2B and C918 groups revealed the consistent results, suggesting that p53 siRNA effectively inhibited the invasion ability of C918 and MUM-2B cells.
Correlation between the expression of p53 in uveal melanoma and the prognosis
All the 88 patients of uveal melanoma were followed up. Among them, 44 patients survived and 44 died. Out of the 88 patients of uveal melanoma, 31 cases revealed low expression of p53 protein, which included 11 death cases, and 57 cases of high expression of p53 protein, which included 33 death cases (Table 3). Kaplan–Meier single-factor analysis showed that different expressions of p53 were of great significance for patient prognosis with a significant P-value of <0.05 (Table 4).
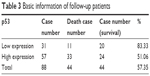  | Table 3 Basic information of follow-up patients |
  | Table 4 Comparison between p53 expression and the operational prognosis |
Discussion
Uveal melanoma has a high incidence rate and is the second most common primary malignant melanoma.8 Because of the high invasion ability of uveal melanoma, most patients have already suffered from the metastasis, when they were diagnosed.9–11 The pathogenesis of uveal melanoma is very complex and still needs further research evidence.12–13 The exact pathogenesis of uveal melanoma is still in the infancy stage of the research. Nonmutant p53 gene is also called wild-type p53 gene, which has two functions: one is to regulate the cell cycle and cellular division, and the other is to promote the cell apoptosis.12,13–17 The content of wild-type p53 protein is low in normal cells with short half-life. It cannot be detected by immunohistochemistry and by other conventional methods. However, the short half-life of mutant p53 protein is relatively long and highly expressed, and is also easy to be detected.18,19
Studies have found that breast cancer patients who showed high expression of p53 protein are usually accompanied with high degree of malignancy and relatively bad operational prognosis.20
The results of the present research have shown that p53 gene was highly expressed in C918 and MUM-2B cells at mRNA and protein levels. In addition, 77.27% of the uveal melanoma tissues showed higher expression of p53 gene. To further analyze the effect of p53 protein in uveal melanoma, gene silencing technique was used to inhibit the expression of p53 protein in uveal melanoma cells. It has been found that after the inhibition of p53 protein, the invasion ability of uveal melanoma cell was also inhibited. Research and correlation analysis of survival prognosis have found that the high expression of p53 and the prognosis of uveal melanoma were negatively correlated.
Out of the 88 patients of uveal melanoma, 31 cases revealed low expression of p53 protein, which included 11 death cases, and 57 cases revealed high expression of p53 protein, which included 33 death cases. Statistical analysis showed that higher expression of p53 protein and the prognosis of uveal melanoma were negatively correlated. Patients with high expression of p53 showed worse prognosis. Therefore, p53 protein can serve as a new biomarker for the diagnosis and prediction of the prognosis of uveal melanoma patients after surgery. The study has a very strong methodology section with newer and modern techniques for the evaluation of p53 gene expression and found that subsequent results have strong clinical correlation.
Limitation
The sample size could be larger, and a long-term longitudinal study should be conducted to prove the results more accurately.
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Eagle RC Jr. The pathology of ocular cancer. Eye (Lond). 2013;27(2):128–136. | ||
McLean IW, Saraiva VS, Burnier MN Jr. Pathological and prognostic features of uveal melanomas. Can J Ophthalmol. 2004;39(4):343–350. | ||
Coupland SE, Lake SL, Zeschnigk M, Damato BE. Molecular pathology of uveal melanoma. Eye (Lond). 2013;27(2):230–242. | ||
Yonekawa Y, Kim IK. Epidemiology and management of uveal melanoma. Hematol Oncol Clin North Am. 2012;26(6):1169–1184. | ||
Graziano SL, Tatum A, Herndon JE 2nd, et al. Use of neuroendocrine markers, p53, and HER2 to predict response to chemotherapy in patients with stage III non-small cell lung cancer: a Cancer and Leukemia Group B study. Lung Cancer. 2001;33(2–3):115–123. | ||
Maniwa Y, Yoshimura M, Obayashi C, et al. Association of p53 gene mutation and telomerase activity in resectable non-small cell lung cancer. Chest. 2001;120(2):589–594. | ||
Thor AD, Yander DW. Prognostic significance of p53 overexpression in node-negative breast carcinoma: preliminary studies support cautious optimism. J Natl Cancer Inst. 1993;85(3):176. | ||
Hurst EA, Harbour JW, Cornelius LA. Ocular melanoma: a review and the relationship to cutaneous melanoma. Arch Dermatol. 2003;13(8):1067–1073. | ||
Bakalian S, Marshall JC, Logan P, et al. Molecular pathways mediating liver metastasis in patients with uveal melanoma. Clin Cancer Res. 2008;14(4):951–956. | ||
Yang TT, Fanq QF, Zhen J, Wu BQ. Molecular basis of the malignant phenotype of the invasive transfer of the human melanoma cell. Recent Adv Ophthalmol. 2000;5:325–327. | ||
Damato B, Groenewald C, MeGalliard J, Wong D. Endoresection of choroidal melanoma. Br J Ophthalmol. 1998;82:213–218. | ||
Eggermont AM. Randomized trials in melanoma: an update. Surg Oncol Clin N Am. 2006;15(2):439–451. | ||
Sato T, Hart F, Yamamoto A. The biology and management of uveal melanoma. Curr Oncol Rep. 2008;10(5):431–438. | ||
Porter PL, Gown AM, Kramp SG, et al. Widespread p53 overexpression in human malignant tumors. An immunohistochemical study using methacarn-fixed, embedded tissue. Am J Pathol. 1992;140:145. | ||
Kastan MB, Berkovich E. p53: a two-faced cancer gene. Nat Cell Biol. 2007;9(5):489–491. | ||
Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2(2):a1107. | ||
Strano S, Dell’Orso S, Di Agostino S, Fontemaggi G, Sacchi A, Blandino G. Mutant p53: an oncogenic transcription factor. Oncogene. 2007;26(15):2212–2219. | ||
Hall PA, Ray A, Lemoine NR, Midgley CA, Krausz T, Lane DP. p53 immunostaining as a marker of malignant disease in diagnostic cytopathology. Lancet. 1991;338(8765):513. | ||
Martinazzi M, Crivelli F, Zampatti C, Martinazzi S. Relationship between p53 expression and other prognostic factors in human breast carcinoma. An immunohistochemical study. Am J Clin Pathol. 1993;100(3):213–217. | ||
Chen W, Li J, Liu C, et al. A functional p53 responsive polymorphism in KITLG, rs4590952, does not affect the risk of breast cancer. Sci Rep. 2014;5(4):63–71. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
