Back to Journals » OncoTargets and Therapy » Volume 12
EPS8 is a Potential Oncogene in Glioblastoma
Received 18 August 2019
Accepted for publication 15 November 2019
Published 2 December 2019 Volume 2019:12 Pages 10523—10534
DOI https://doi.org/10.2147/OTT.S227739
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Gang Yang,1,2 Yong-bin Lu,3 Quan-lin Guan2,4
1Department of Neurosurgery, The First Hospital of Lanzhou University, Lanzhou, Gansu 730000, People’s Republic of China; 2The First Clinical Medical College of Lanzhou University, Lanzhou, Gansu 730000, People’s Republic of China; 3Department of Technology, The First Hospital of Lanzhou University, Lanzhou, Gansu 730000, People’s Republic of China; 4Department of Oncology Surgery, The First Hospital of Lanzhou University, Lanzhou, Gansu 730000, People’s Republic of China
Correspondence: Quan-lin Guan
Department of Oncology Surgery, The First Hospital of Lanzhou University, Donggang West Road, Chengguan District, Lanzhou, Gansu Province 730000, People’s Republic of China
Tel|Fax +86 931 861 9797
Email [email protected]
Purpose: In this study, we investigated the expression and function of Epidermal growth factor receptor kinase substrate 8 (EPS8) in glioblastoma (GBM), and further explored the underlying mechanisms that regulate it.
Patients and methods: The expression and potential mechanisms of EPS8 in GBM were evaluated through multiple online public databases. The expression level EPS8 in GBM tissues and cell lines were detected by immunohistochemical staining and Western blot. Then, the prognosis of EPS8 and GBM patients were analyzed. Loss-of-function experiments were conducted to determine the role of EPS8 for the biological behavior of GBM cells. In addition, the tumorigenic ability of nude mice was tested in vivo.
Results: EPS8 is highly expressed in GBM tissues and indicates poor patient prognosis. In cell experiments, EPS8 can promote the proliferation, migration and invasion of GBM cells. In vivo, EPS8 promotes tumor formation in nude mice. EPS8 can activate the PI3K/Akt signaling pathway to function.
Conclusion: EP8S plays a role in the development of GBM and may be a potential therapeutic target for GBM.
Keywords: GBM, EPS8, PI3K/Akt pathway
Introduction
Gliomas are the most common form of tumors in the central nervous system, accounting for approximately 80% of malignant brain tumors.1 According to the World Health Organization (WHO), gliomas can be classified into four groups (grades I–IV). Among them, glioblastomas (GBM; WHO grade IV) have the highest incidence, accounting for 46.1% of all gliomas.2 Gliomas are biologically characterized by invasive and diffuse growth. Therefore, it is difficult to completely remove GBM by surgery, resulting in frequent postoperative relapse. Despite the rapid development of comprehensive anti-tumor models in recent years, effective therapies for treating gliomas remain unsatisfactory. Diverse genetic, epigenetic, and developmental programs drive glioblastoma research, an incurable and poorly understood tumor, but their precise characterization remains challenging.3 Therefore, the present research focused on exploring GBM pathogenesis at the molecular level to identify therapeutic targets.
Epidermal growth factor receptor kinase substrate 8 (EPS8) is a part of the epidermal growth factor receptor (EGFR) signaling pathway.4,5 EPS8 can act as an adaptor protein to bind several chaperone proteins, forming complexes to regulate multiple signaling pathways.6 Physiologically, EPS8 is involved in actin remodeling mediated by growth factors.7 Additionally, EPS8 is closely associated with the formation of cell microvilli, cell adhesion, motility, and angiogenesis.8–10 EPS8 has been identified as an oncogene in malignant solid tumors.11 Anomalies in EPS8 expression are involved in malignant progression of solid tumor cells, such as in lung, breast, and pancreatic cancer, as well as, in squamous cell carcinomas.12–15 However, studies of EPS8 in GBM are relatively rare, and its regulation mechanisms are unclear. In this study, we investigated EPS8 expression and function in GBM, and further explored underlying regulatory mechanisms.
Materials and Methods
Bioinformatics Analysis
A variety of online databases were used to predict the expression and prognostic significance of EPS8 in GBM. Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn) was first used to analyze differential expression of EPS8 mRNA in GBM samples and normal samples. In particular, GEPIA was used to analyze RNA sequencing data from 9736 tumor samples and 8587 normal samples from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) programs. In addition, the Chinese Glioma Genome Atlas (CGGA; http://www.cgga.org.cn) was used to analyze EPS8 expression levels in gliomas of different grades to predict their prognostic effect in GBM patients. Notably, the CGGA contains over 2000 samples from Chinese glioma patients, which were used to analyze mRNA expression profiles and prognosis in glioma patients. Finally, Gene Set Enrichment Analysis (GSEA) was used for signal pathway enrichment analysis, aiming to identify EPS8-mediated molecular pathways in GBM.
Tissue Samples
Paraffin-embedded tissues of GBM patients (N = 98) who underwent surgical resection at First Hospital of Lanzhou University between January 2005 and December 2014 were collected. All 98 GBM patients were pathologically diagnosed and had complete clinical data. The mean age of the patients was 46.2 ± 12.1 years (range 35–72 years), with 68 males and 30 females. In addition, three pairs of fresh GBM tissues and adjacent non-tumor tissues were collected for Western blot analysis. None of the enrolled GBM patients received chemotherapy, radiation, or biotherapy before surgery. All patients signed written informed consent. The study was approved by the Ethics Committee of the First Hospital of Lanzhou University.
Cell Culture and Transfection
GBM cell lines, U251, U87MG, SHG44, and A172 were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). The cells were maintained in RPMI 1640 medium (Invitrogen, Shanghai, China) containing 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 0.1 mg/mL streptomycin at 37°C with 5% CO2. Culture medium was changed every other day.
Two different short hairpin RNA (shRNA) target sequences were designed based on the design principles of RNA interference sequences. The target sequences of EPS8-shRNA-1 and EPS8-shRNA-2 were GGGAGCCACAATGGAACAAGA and GCGAGAGTCTATAGCCAAATC, respectively. The lentiviruses with these target sequences were commercially prepared and packaged (GeneChem, Shanghai, China). Selected GBM cells were infected with lentivirus according to the manufacturer’s instructions, followed by puromycin selection (5 μg/mL). EPS8 knockdown efficiency was assessed by Western blot.
Immunohistochemistry and Evaluation
The streptavidin-peroxidase method was used for immunohistochemistry (IHC). Paraffin-embedded tissue was cut into 5-μm-thick sections, followed by dewaxing and dehydration. Endogenous biotin was blocked with a 3% methanol in H2O2 solution, and then a 10% BSA solution was used to block non-specific binding. Subsequently, the sections were incubated with primary antibody (Rabbit Anti-EPS8 antibody, 1:100; Abcam, Cambridge, MA) at 4°C overnight. The sections were then washed with phosphate buffer saline (PBS), and then incubated with secondary antibody (Zhongshan Golden Bridge Biotechnology, Beijing, China) for 30 mins at 30°C. Finally, 3,3ʹ-diaminobenzidine tetrahydrochloride was used for visualization, and hematoxylin was added for counterstaining.
All IHC sections were independently scored by two pathologists. The intensity of positive staining in cells was divided into four grades: 0 points without staining; 1 point for lightly brown; 2 points for moderately brown; and 3 points for strongly brown. The percentage of positively stained cells was also categorized into four grades: 0 for no staining; 1 for <25%; 2 for 25–75%; and 3 for >75%. Subsequently, the two scores were multiplied to obtain the final score. A score over 5 indicated a high EPS8 expression, and a score less than or equal to 5 suggested low EPS8 expression.
Western Blot
Cells were harvested to extract total protein using RIPA lysis buffer (Beyotime Institute of Biotechnology, Beijing, China) on ice. Tissue samples were thoroughly ground before adding RIPA buffer. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate protein samples, which were later transferred to polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After the membranes were blocked with 5% milk, they were incubated with primary antibody (Anti-EPS8 antibody, 1:1500; Abcam) overnight at 4°C. After incubation with the secondary antibody at room temperature for 1 h, an enhanced chemiluminescent (ECL) kit (Biorobot, Shanghai, China) was used for visualization.
Cell Proliferation Assay
Cell counting kit-8 (CCK-8; Dojindo, Kumamoto, Japan) was used to evaluate the proliferative capability of GBM cells. Briefly, cells were seeded into 96-well plates at a density of 2000 cells/per well. After incubation for 0, 24, 48, 72 and 96 hrs, cell proliferation was determined by adding the CCK-8 solution according to the manufacturer’s instructions. A microplate reader was used to measure optical density at 450 nm (Bio-Rad Laboratories, Hercules, CA, USA). The assay was conducted in triplicate.
Cell Migration and Invasion Assay
Transwell chambers were used to conduct invasion and migration assays (Corning, NY, USA). Pre-coated Matrigel Transwell chambers (BD Biosciences, Franklin Lakes, USA) were used to assess invasion ability, while Transwell chambers without Matrigel were utilized to determine migration capacity. First, the cells were starved in serum-free medium for 24 hrs. The lower chamber was filled with medium containing 20% FBS as a chemotactic agent. After incubation for 24 hrs, cells that migrated or invaded the bottom of the membrane were fixed with 75% methanol for 30 mins, and then stained with 0.5% crystal violet for 1 h. Finally, the proportion of migrated or invaded cells was calculated.
Wound-Healing Assay
A wound-healing assay was employed to assess the migration ability of GBM cells. Approximately 2 × 105 cells were seeded into 6-well plates and incubated until cell confluency reached 100%. Then, a plastic pipette tip was used to gently scrape the monolayer of cells. The cells were washed three times with PBS and further incubated with serum-free medium at 37°C with 5% CO2. Cells were observed and images were captured with a microscope at 0 and 24 hrs, followed by calculation of wound-healing rate.
Tumorigenicity Assay in Nude Mice
Experiments with xenograft tumors were performed using 6–7 weeks old BALB/c male nude mice (Charles River; Beijing, China). Nude mice were divided into negative control (NC) group and EPS8-shRNA-2 group. Next, U87MG cells in logarithmic growth phase in the NC and EPS8-shRNA groups were collected, and a cell suspension of 5 × 107 cells/mL was prepared. Then, 0.2 mL of cells was inoculated subcutaneously into nude mice. A clear bulge can be seen at the injection site to indicate successful vaccination. After injection, the volume of subcutaneous tumors in nude mice was measured every 5 days. Nude mice were sacrificed 25 days after inoculation, and tumors were removed for analysis. Animal experiments were conducted under the approval of the Animal Ethics Committee of The First Hospital of Lanzhou University and were performed in accordance with the Guide for the Care and Use of Laboratory animals published by the US National Institutes of Health.
Statistical Analysis
Data are shown as mean ± standard deviation (SD). Unpaired t-tests were used to determine differences between groups. Correlation between the clinicopathological parameters of patients with glioblastoma and EPS8 was evaluated by the Chi-square test. Kaplan–Meier and log-rank tests were used to assess the correlation between EPS8 expression and survival of GBM patients. Overall survival (OS) and disease-free survival (DFS) were defined as primary outcome measures. SPSS 19.0 software (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 7 (GraphPad Software Inc., San Diego, CA) were used for statistical analysis. A P value <0.05 was considered as statistically significant.
Results
Bioinformatics Prediction
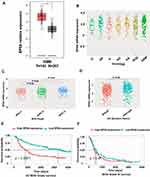
First, EPS8 expression in GBM was predicted through various databases. Based on the TCGA database, GEPIA analysis indicated that EPS8 mRNA expression was significantly higher in GBM samples than in normal samples (Figure 1A). EPS8 expression in glioma and GBM was further predicted online in the CGGA database. As a result, EPS8 expression was significantly increased in GBM compared with other pathological types of glioma (Figure 1B). According to the WHO classification of gliomas, EPS8 expression was significantly higher in GBM than in other subtypes of glioma (Figure 1C). In addition, higher EPS8 expression was detected in gliomas with wild-type isocitrate dehydrogenase (IDH) (Figure 1D). Based on prognostic data of glioma patients from the CGGA database, we further analyzed the association between EPS8 and prognosis of patients with glioma. Consequently, glioma patients with high expression of EPS8 had a poorer prognosis than those with low expression (Figure 1E). Although the prognosis of GBM patients with low expression of EPS8 was slightly better than those with high expression, the difference was not statistically significant (Figure 1F).
Correlation Between EPS8 and GBM
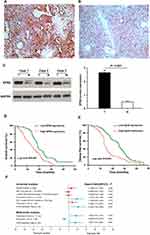
To investigate the importance of EPS8 in GBM progression, IHC was used to examine its expression in GBM tissues and corresponding adjacent tissues, revealing primarily cytoplasmic localization of EPS8 protein (Figure 2A and B). High expression rate of EPS8 found in tumor tissues (57.1%, 56/98) was significantly more prominent than that of corresponding non-tumor tissues (37.8%, 37/98). We also quantified EPS8 expression by Western blot in three pairs of GBM and corresponding adjacent non-tumor tissues. As shown in Figure 2C, EPS8 expression in GBM tissues was significantly higher than in non-tumor tissues. Subsequently, the analysis of the association between the clinicopathological features of patients with GBM and EPS8 expression was performed. The results are summarized in Table 1, where high expression of EPS8 was closely associated with tumor size and Karnofsky Performance Score (KPS) in GBM patients (P < 0.05), but not with gender, age, or tumor location. Intriguingly, our results showed that EPS8 expression was not related to IDH gene mutations in GBM patients, which was not consistent with previous bioinformatics predictions.
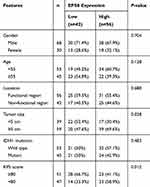 |
Table 1 Correlation Between the EPS8 Expression and Clinicopathological Features (n = 98) |
The effects of EPS8 expression on DFS and OS in patients with GBM were assessed by Kaplan-Meier survival (KPS) analysis. Log-rank test results showed that high EPS8 expression was associated with poor OS and DFS (Figure 2D and E). To further evaluate whether EPS8 expression was an independent prognostic factor for GBM, both univariate and multivariate Cox regression analysis models were used to assess its prognostic significance. Univariate Cox regression analysis revealed that high expression of EPS8 was an important factor for OS in GBM patients (Table 2 and Figure 2F). Multivariate Cox regression analysis further showed that, besides tumor size (HR = 1.658, 95% CI: 1.051–2.616, P < 0.05) and KPS score (HR = 0.468, 95% CI: 0.303–0.723, P < 0.05), high expression of EPS8 was also an independent prognostic factor for OS (HR = 1.651, 95% CI: 1.014–2.689, P < 0.05) (Table 2 and Figure 2E). Therefore, we further investigated the biological functions and molecular mechanisms of EPS8 in GBM progression.
 |
Table 2 Univariate and Multivariable Analyses of Overall Survival |
EPS8 Promoted Proliferation, Migration, and Invasion of GBM Cells
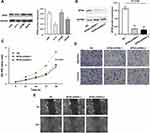
EPS8 protein expression in four GBM cell lines was detected by Western blot. As shown in Figure 3A, EPS8 expression was the highest in U87MG; therefore, the U87MG cell line was used for subsequent functional validation. Lentiviral-mediated shRNA was used to construct a stable cell line with EPS8 knockdown. EPS8 expression was significantly down-regulated following infection with two different shRNAs targeting EPS8, compared with that of untreated U87MG cells (NC group) (Figure 3B).
CCK-8 assays were further used to assess the effect of EPS8 on U87MG proliferation. The knockdown of EPS8 inhibited cell proliferation in U87MG (P < 0.05, Figure 3C). Transwell chambers were used to analyze the effects of EPS8 knockdown on cell migration and invasion, showing decreased cell migration and invasion in U87MG cells (P < 0.05, Figure 3D). Moreover, wound healing assays were performed to investigate the effects of EPS8 knockdown on GBM cell migration, showing significantly decreased U87MG cell motility (P < 0.05, Figure 3E).
High Expression of EPS8 Activated PI3K/Akt Signaling Pathway
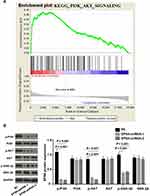
We further explored the molecular mechanisms of EPS8 in GBM progression. Aberrant activation of the phosphatidylinositol 3-kinase/protein kinase B (PI3K/Akt) signaling pathway is known to promote tumor progression. Previous GSEA analysis suggested that high EPS8 expression was mainly enriched in the PI3K/Akt signaling pathway (Figure 4A). Therefore, the phosphorylation status of PI3K, Akt, and glycogen synthase 3 beta (GSK-3β) proteins after EPS8 knockdown was assessed by Western blot to determine whether the observed phenomenon was caused by decreased PI3K/Akt signaling transduction. Total protein expression of PI3K, Akt, and GSK-3β was not significantly changed in the two EPS8-shRNA U87MG groups compared to NC cells. However, EPS8 knockdown significantly inhibited levels of phosphorylated PI3K, Akt, and GSK-3β in U87MG cells (Figure 4B).
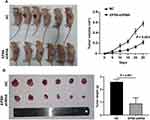
EPS8 Promotes Tumor Growth in vivo
To explore the effect of EPS8 on GBM tumor growth in vivo, U87MG cells of the NC and EPS8-shRNA groups were inoculated subcutaneously into BALB/c nude mice. After successful inoculation of U87MG cells, the volume of subcutaneous tumors in nude mice was measured every 5 days. The results showed that both groups of nude mice had tumor formation under the skin. The subcutaneous tumor growth rate in nude mice of the EPS8 group was slower than that of the NC group (Figure 5A). After 25 days, we sacrificed all of the mice and dissected the tumors. We found that the tumor weight of the EPS8 group of mice was significantly lower than that of the NC group (Figure 5B). Therefore, EPS8 knockdown inhibited tumor growth in nude mice.
Discussion
EPS8 is a substrate for receptor tyrosine kinases, and contains an Src homology 3 (SH3) domain, playing an important role in promoting mitotic signaling.16 The EPS8 gene is located on chromosome 12q23-q24 in humans.17 EPS8 protein includes a specific N-terminal phospho-tyrosine-binding domain, a central SH3 domain, and a C-terminal “effector region”.18 The latter region regulates intracellular EPS8 localization and is sufficient to activate GTPase and Rac to remodel the actin cytoskeleton.18 EPS8 has been confirmed to enhance the reactivity of epidermal growth factor (EGF), thereby promoting EGF binding to its receptor, EGFR, to activate mitotic signal transmission.19 Studies have shown that EPS8 forms a Rac-specific guanine nucleotide exchange factor complex with Abi-1 and Sos-1, and that motility and invasiveness of fibrosarcoma cells could be inhibited by suppressing the formation of this complex.20,21 A number of studies have also indicated that abnormal EPS8 expression is closely related to biological activity in several types of cancer. Maa et al have reported that EPS8 advances motility and growth of cells in colon cancer by enhancing activity and expression of focal adhesion kinase.22 Welsch et al found that EPS8 expression is significantly increased in pancreatic cancer, and localizes to F-actin filaments, filopodia, and the tip of the cell advancing front. They also observed that EPS8 knockdown altered cell morphology and the actin-based cytoskeletal structure, attenuating protrusion formation and cell-cell junctions.23 Moreover, EPS8 is closely associated with growth and metastasis of malignant tumors, such as cervical cancer, oral squamous cell carcinoma, and ovarian cancer.24–26
Studies concerning EPS8 in GBM are rare. A Chinese study reported that the rate of EPS8 overexpression in high-grade glioma tissues was significantly higher than that in normal brain tissues by IHC, and plays a tumor-promoting role through extracellular receptor kinase (ERK) and Akt/β-catenin pathways.27 Cattaneo et al used small interfering RNA (siRNA) to silence EPS8 expression in GBM cell lines, and subsequent in vitro assays showed that EPS8 silencing effectively inhibited GBM cell migration and invasion.28 In this study, our research direction and hypotheses were first established through bioinformatics. Differential expression of EPS8 was further confirmed in clinical samples comparing GBM and non-tumor tissues, which is consistent with results reported by Ding et al.27 Statistical analysis showed that EPS8 was closely related to the clinicopathological features, including tumor size and KPS score in GBM patients. A clinical predictive model revealed that GBM patients with high EPS8 expression had shorter survival time, and that EPS8 is an independent risk factor for prognosis in GBM patients. In vitro assays were critical to this study. Lentiviral-mediated shRNA was used to effectively silence EPS8 expression in GBM cells. As a result, EPS8 knockdown suppressed cell proliferation, migration, and invasion of U87MG cells, which was similar to results reported by Cattaneo et al.28 In vivo experiments showed that EPS8 can promote tumor growth. The above findings further support that EPS8 promotes GBM progression.
In this study, we also focused on the mechanism by which EPS8 played in GBM progression. Outcomes from GSEA analysis indicated a close association between EPS8 and PI3K/Akt signaling. The PI3K/Akt signaling pathway plays an important role in GBM progression. Zheng et al found that interleukin 17A (IL-17A) promotes migration and invasion of GBM by activating the PI3K/Akt signal transduction.29 Using an open-access data set in the TCGA database containing 608 GBM samples, Park et al analyzed and validated that the PI3K/Akt pathway is critical for predicting the prognosis of GBM patients.30 In addition, EPS8 has been implicated in regulating proliferation, apoptosis, and chemosensitivity of BCR-ABL-positive cells via the PI3K/Akt signaling pathway in chronic myeloid leukemia.31 Therefore, we further examined changes in the expression of related molecules in the PI3K/Akt signaling pathway. Western blot confirmed that EPS8 promoted tumor formation by activating the PI3K/Akt pathway in GBM progression.
Conclusion
In conclusion, our study provides new evidence for clinical and biological significance of EPS8 in GBM. Our data showed a high expression of EPS8 in GBM, which has been identified as a predictive marker for the prognosis of GBM patients. Notably, we have found that EPS8 promotes malignant biological behavior of GBM by activating the PI3K/Akt signaling pathway. However, further study of the mechanisms involved is needed. In addition, patient-derived xenografts (PDX) can provide more evidence for this study. We conclude that EPS8 may be a potential therapeutic target for GBM.
Acknowledgment
This project was supported in part by grants from The Natural Science Foundation of Gansu Province (NO. 18JR3RA348) and The Health Research Plan Management Project of Gansu Province (NO. GWGL2014-49 and GSWSKY2016-21).
Author Contributions
Gang Yang and Quan-lin Guan conceived this study and took responsibility for the quality of the data. Yong-bin Lu acquired data and played an important role in interpreting the results. Gang Yang performed the data analysis and wrote the manuscript. Quan-lin Guan played an important role in modifying the paper. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas – implications for classification and therapy. Nat Rev Clin Oncol. 2017;14(7):434–452. doi:10.1038/nrclinonc.2016.204
2. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi:10.1007/s00401-016-1545-1
3. Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849. doi:10.1016/j.cell.2019.06.024
4. Offenhäuser N, Castelletti D, Mapelli L, et al. Increased ethanol resistance and consumption in Eps8 knockout mice correlates with altered actin dynamics. Cell. 2006;127(1):213–226. doi:10.1016/j.cell.2006.09.011
5. Lanzetti L, Rybin V, Malabarba MG, et al. The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature. 2000;408(6810):374–377. doi:10.1038/35042605
6. Welsch T, Younsi A, Disanza A, et al. Eps8 is recruited to lysosomes and subjected to chaperone-mediated autophagy in cancer cells. Exp Cell Res. 2010;316(12):1914–1924. doi:10.1016/j.yexcr.2010.02.020
7. Giampietro C, Disanza A, Bravi L, et al. The actin-binding protein EPS8 binds VE-cadherin and modulates YAP localization and signaling. J Cell Biol. 2015;211(6):1177–1192. doi:10.1083/jcb.201501089
8. Postema MM, Grega-Larson NE, Neininger AC, Tyska MJ. IRTKS (BAIAP2L1) elongates epithelial microvilli using EPS8-dependent and independent mechanisms. Curr Biol. 2018;28(18):2876–2888. doi:10.1016/j.cub.2018.07.022
9. Stamatakou E, Marzo A, Gibb A, Salinas PC. Activity-dependent spine morphogenesis: a role for the actin-capping protein Eps8. J Neurosci. 2013;33(6):2661–2670. doi:10.1523/JNEUROSCI.0998-12.2013
10. Cappellini E, Vanetti C, Vicentini LM, Cattaneo MG. Silencing of Eps8 inhibits in vitro angiogenesis. Life Sci. 2015;131:30–36. doi:10.1016/j.lfs.2015.03.018
11. Wang Q, Yong L. Eps8, a therapeutic potential target for cancer. Hum Pathol. 2018;82:300–301. doi:10.1016/j.humpath.2018.04.037
12. Wen Q, Jiao X, Kuang F, et al. FoxO3a inhibiting expression of EPS8 to prevent progression of NSCLC: a new negative loop of EGFR signaling. EBioMedicine. 2019;40:198–209. doi:10.1016/j.ebiom.2019.01.053
13. Ohshima K, Hatakeyama K, Kanto K, et al. Comparative proteomic analysis identifies exosomal Eps8 protein as a potential metastatic biomarker for pancreatic cancer. Oncol Rep. 2019;41(2):1019–1034. doi:10.3892/or.2018.6869
14. Chen C, Liang Z, Huang W, et al. Eps8 regulates cellular proliferation and migration of breast cancer. Int J Oncol. 2015;46(1):205–214. doi:10.3892/ijo.2014.2710
15. Schoenherr C, Serrels B, Proby C, et al. Eps8 controls Src- and FAK-dependent phenotypes in squamous carcinoma cells. J Cell Sci. 2014;127(Pt 24):5303–5316. doi:10.1242/jcs.157560
16. Biesova Z, Piccoli C, Wong WT. Isolation and characterization of e3B1, an eps8 binding protein that regulates cell growth. Oncogene. 1997;14(2):233–241. doi:10.1038/sj.onc.1200822
17. Wong WT, Carlomagno F, Druck T, et al. Evolutionary conservation of the EPS8 gene and its mapping to human chromosome 12q23-q24. Oncogene. 1994;9(10):3057–3061.
18. Di Fiore PP, Scita G. Eps8 in the midst of GTPases. Int J Biochem Cell Biol. 2002;34(10):1178–1183. doi:10.1016/S1357-2725(02)00064-X
19. Matoskova B, Wong WT, Salcini AE, Pelicci PG, Di Fiore PP. Constitutive phosphorylation of eps8 in tumor cell lines: relevance to malignant transformation. Mol Cell Biol. 1995;15(7):3805–3812. doi:10.1128/MCB.15.7.3805
20. Funato Y, Terabayashi T, Suenaga N, Seiki M, Takenawa T, Miki H. IRSp53/Eps8 complex is important for positive regulation of Rac and cancer cell motility/invasiveness. Cancer Res. 2004;64(15):5237–5244. doi:10.1158/0008-5472.CAN-04-0327
21. Disanza A, Carlier MF, Stradal TE, et al. Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat Cell Biol. 2004;6(12):1180–1188. doi:10.1038/ncb1199
22. Maa MC, Lee JC, Chen YJ, et al. Eps8 facilitates cellular growth and motility of colon cancer cells by increasing the expression and activity of focal adhesion kinase. J Biol Chem. 2007;282(27):19399–19409. doi:10.1074/jbc.M610280200
23. Welsch T, Endlich K, Giese T, Büchler MW, Schmidt J. Eps8 is increased in pancreatic cancer and required for dynamic actin-based cell protrusions and intercellular cytoskeletal organization. Cancer Lett. 2007;255(2):205–218. doi:10.1016/j.canlet.2007.04.008
24. Chen YJ, Shen MR, Chen YJ, Maa MC, Leu TH. Eps8 decreases chemosensitivity and affects survival of cervical cancer patients. Mol Cancer Ther. 2008;7(6):1376–1385. doi:10.1158/1535-7163.MCT-07-2388
25. Yap LF, Jenei V, Robinson CM, et al. Upregulation of Eps8 in oral squamous cell carcinoma promotes cell migration and invasion through integrin-dependent Rac1 activation. Oncogene. 2009;28(27):2524–2534. doi:10.1038/onc.2009.105
26. Chen H, Wu X, Pan ZK, Huang S. Integrity of SOS1/EPS8/ABI1 tri-complex determines ovarian cancer metastasis. Cancer Res. 2010;70(23):9979–9990. doi:10.1158/0008-5472.CAN-10-2394
27. Ding X, Zhou F, Wang F, et al. Eps8 promotes cellular growth of human malignant gliomas. Oncol Rep. 2013;29(2):697–703. doi:10.3892/or.2012.2160
28. Cattaneo MG, Cappellini E, Vicentini LM. Silencing of Eps8 blocks migration and invasion in human glioblastoma cell lines. Exp Cell Res. 2012;318(15):1901–1912. doi:10.1016/j.yexcr.2012.05.010
29. Zheng Q, Diao S, Wang Q, et al. IL-17A promotes cell migration and invasion of glioblastoma cells via activation of PI3K/AKT signalling pathway. J Cell Mol Med. 2019;23(1):357–369. doi:10.1111/jcmm.13938
30. Park AK, Kim P, Ballester LY, Esquenazi Y, Zhao Z. Subtype-specific signaling pathways and genomic aberrations associated with prognosis of glioblastoma. Neuro Oncol. 2019;21(1):59–70. doi:10.1093/neuonc/noy120
31. Huang R, Liu H, Chen Y, et al. EPS8 regulates proliferation, apoptosis and chemosensitivity in BCR-ABL positive cells via the BCR-ABL/PI3K/AKT/mTOR pathway. Oncol Rep. 2018;39(1):119–128. doi:10.3892/or.2017.6102
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.