Back to Journals » Infection and Drug Resistance » Volume 14
Epidemiology of Biofilm Producing Acinetobacter baumannii Nosocomial Isolates from a Tertiary Care Hospital in Egypt: A Cross-Sectional Study
Authors Asaad AM , Ansari S , Ajlan SE, Awad SM
Received 13 May 2020
Accepted for publication 4 February 2021
Published 23 February 2021 Volume 2021:14 Pages 709—717
DOI https://doi.org/10.2147/IDR.S261939
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sahil Khanna
Ahmed Morad Asaad,1 Shamshul Ansari,2 Soma Elsayed Ajlan,3 Samah Mohammed Awad4
1Department of Medical Microbiology and Immunology, Faculty of Medicine, Zagazig University, Zagazig, Egypt; 2Department of Microbiology and Immunology, Chitwan Medical College School of Medicine, Bharatpur, 44200, Nepal; 3Department of Medical Microbiology and Immunology, Faculty of Medicine, Menoufia University, Menoufia, Egypt; 4Department of Clinical Microbiology and Immunology, Molecular Microbiology in Liver and GIT, National Liver Institute, Menoufia, Egypt
Correspondence: Shamshul Ansari
Department of Microbiology, Chitwan Medical College School of Medicine, Bharatpur, 44200, Chitwan, Nepal
Tel +977-9840656933
Email [email protected]
Objective: This cross-sectional study aims to determine the prevalence and associated risk factors of biofilm-producing A. baumannii nosocomial isolates from a tertiary care hospital, as well as to investigate any possible association of biofilm formation with the distribution of biofilm-related genotypes and antibiotic resistance phenotypes.
Methods: A total of 94 non-duplicate A. baumannii nosocomial isolates were identified, their biofilm formation was quantitatively detected using the modified microtiter plate assay, and their susceptibilities to different antibiotics were determined using the breakpoint method. Isolates were then subjected to PCR assays targeting bap, ompA and blaPER-1 genes.
Results: The majority (70.1%) of isolates were biofilm producers. The most prevalent biofilm gene was ompA (63.8%), followed by bap (13.8%) and blaPER-1 (10.6%). The presence of multi- and extensive-drug resistance (MDR and XDR) was significantly associated with biofilm producers (p = 0.017 and 0.002, respectively). The length of hospital stay (aOR= 0.023), the presence of ompA gene (aOR = 0.286) or bap gene (aOR = 0.346), ampicillin/sulbactam resistance (aOR = 1), and the presence of MDR (aOR = − 0.329) or XDR (aOR = − 0.252) were considered significant risk factors associated with biofilm-producing isolates.
Conclusion: The high prevalence of biofilm-producing MDR and XDR nosocomial isolates in this study is worrisome and alarming. Characterization of risk factors could help control the continuous selection and transfer of this serious A. baumannii phenotype inside hospitals and improve the quality of patients’ care.
Keywords: Acinetobacter, biofilm, MDR, XDR, nosocomial infection
Introduction
A. baumannii is still considered a serious nosocomial pathogen as it accounts for a wide range of healthcare-associated infections, including bacteraemia, urinary tract infection, secondary meningitis, skin and soft tissues infection, and nosocomial and ventilator-associated pneumonia, especially in patients admitted to intensive care and burn units.1–3 These infections are usually associated with a high mortality rate of up to 26% for hospitalized patients4 to up to 43% for intensive care unit (ICU) patients.5 A. baumannii is the first in the critical priority list of pathogens that poses the greatest threat to human health according to the World Health Organization.6 In the last few years, the scenario is more dramatic with a continuous surge of multi-, extensive- and pan-drug resistant (MDR, XDR, and PDR) A. baumannii nosocomial isolates, some of which are even resistant to tigecycline and colistin, the last resort drugs in therapeutic protocols.
Worldwide, the successful model of A. baumannii as an endemic pathogen in healthcare facilities has been attributed to several factors, such as its remarkable intrinsic and acquired resistance to multiple antimicrobial classes, including penicillins, extended-spectrum cephalosporins, fluoroquinolones and carbapenems,1,2 its ability to produce biofilm and persist on biotic and abiotic surfaces such as environmental surfaces and medical devices,3 and its high intake ability of foreign genetic elements to survive under harsh conditions and antibiotic treatment.1,7
The ability of A. baumannii to form biofilm is primarily considered an effective strategy to increase the bacterial survival, adherence to mucosal surfaces, dormancy in deep biofilm layers, and persistence in hospital environment under stress conditions.1,7,8 Several virulence factors are involved in the biofilm formation of A. baumannii such as the biofilm-associated protein (Bap) encoded by the bap gene, the outer membrane protein A (OmpA) encoded by the ompA gene,9 chaperon-usher pilus (Csu), extracellular exopolysaccharide (EPS), two-component system (BfmS/BfmR), poly-β-(1,6)-N-acetyl glucosamine (PNAG) and quorum sensing system.8–11 Besides, Lee et al7 have shown that the ability of clinical isolates of A. baumannii to form biofilm and to adhere to respiratory epithelial cells is enhanced by the presence and expression of the blaPER-1 gene.
Providing a new insight into the better understanding of the possible association between biofilm formation of A. baumannii nosocomial isolates and their antibiotic resistance phenotypes could be helpful for improving infection control procedures in healthcare facilities. However, the findings of previous clinical and epidemiological studies are still scarce and controversial.8–16 Therefore, this study aims to determine the prevalence and associated risk factors of biofilm-producing A. baumannii nosocomial isolates from an Egyptian tertiary care hospital, as well as to investigate any possible association of biofilm formation with the distribution of biofilm-related genotypes and antibiotic resistance phenotypes.
Materials and Methods
This cross-sectional observational study was conducted from August 2018 to July 2019 at the Menoufia University Hospitals (MUHs), a 760-bed tertiary care hospital in Shebeen El-Koum, a city in Egypt. The study was conducted according to the international guidelines of Strengthening the Reporting for Observational Studies in Epidemiology (STROBE).17 During the study period, all A. baumannii nosocomial isolates were collected from adult patients (one isolate from each patient) who were hospitalized for ≥48 hours and classified according to the Centres for Disease Control and Prevention/National Healthcare Safety Network (CDC/NHSN) criteria.18
Demographic and Medical Details
Demographic and medical data of the hospitalized patients were obtained using a questionnaire, which included age, gender, type of (system involved for) infection (urinary tract infection, respiratory tract infection, skin and soft tissue infection, and primary bacteremia), ward admission (medical, surgical, burn unit and ICU), presence of comorbid conditions such as diabetes mellitus, hypertension, chronic pulmonary disease, and chronic heart disease, use of invasive procedures (central or peripheral venous catheter, urinary catheter and ventilator), length of stay, and history of previous antibiotic therapy.
Laboratory Processing
The clinical specimens were collected from all patients and were sent to the microbiology laboratory department of the NLI in Menoufia for further processing. All A. baumannii isolates were identified using conventional biochemical tests,19 the API 20NE (bioMerieux, Marcy-l’Etoile, France) and the VITEK 2 compact system (AST-GN90 susceptibility cards, bioMerieux), according to the manufacturers’ instructions. Antimicrobial susceptibility testing was performed with the broth microdilution method for determination of the minimum inhibitory concentration (MIC) breakpoints, using the VITEK 2 compact system, and the E-test minimum inhibitory concentration method, using E-test strips (AB Biodisk, Solna, Sweden) on Mueller Hinton agar plates using Clinical and Laboratory Standards Institute (CLSI) guidelines.19 The antibiotics tested included ceftazidime, cefotaxime, cefepime, ampicillin/sulbactam, piperacillin/tazobactam, piperacillin, amikacin, gentamycin, tobramycin, ciprofloxacin, gatifloxacin, trimethoprim/sulfamethoxazole, imipenem, meropenem, tigecycline, and colistin. Pseudomonas aeruginosa ATCC 27,853 and Escherichia coli ATCC 25,922 were used as quality control during antibiotic resistance testing.20 To determine the susceptibility to antimicrobial agents, MIC breakpoints from the CLSI guidelines20 were used as interpretative standards except for the tigecycline breakpoints, which were obtained from the US Food and Drug Administration (FDA).21 The MDR was defined as resistance to ≥3 antimicrobial classes (cephalosporins, β-lactam/β-lactamase inhibitor combinations, aminoglycosides, antipseudomonal carbapenems, and fluoroquinolones), and the XDR as resistance to all antibiotics, except colistin or tigecycline.21
Quantitative Biofilm Formation Assay
Biofilm formation was quantitatively detected using the modified microtiter plate assay as previously described12 with some modification. Briefly, after overnight incubation of all isolates inoculated into Tryptic-Soy Broth (TSB, Merck, Germany) containing 0.5% glucose at 37°C, cultures were diluted 1:40 in TSB containing 0.5% glucose. Then, 100 ųl of 108 CFU/mL of A. baumannii and an equal volume of TSB containing 0.5% glucose were added to each well of 96-well polystyrene microtiter plates (Nunc, Denmark), and incubated at 37°C for 48 hours. The well containing 200 µL of TSB with 0.5% glucose was considered as a negative control. Wells were slowly washed three times with Phosphate Buffered Saline (PBS; pH 7.2; Invitrogen, USA), fixed by methyl alcohol (Merck, Germany) for 20 minutes, dried at 20–25°C, and then stained by 0.1% safranin (Merck, Germany). Finally, the optical density (OD) of each well was measured at 490 nm. Optical density cut-off (ODc) was defined as 3X standard deviation above mean OD of the negative control. The isolates were divided into four groups according to their OD results including non biofilm producer (OD ≤ ODc), weak biofilm producer (ODc < OD ≤ 2× ODc), moderate biofilm producer (2× ODc < OD ≤ 4 × ODc), and strong biofilm producer (4× ODc < OD).
PCR Detection of Biofilm-Related Genes
All isolates were subjected to PCR assays to detect bap, ompA and blaPER-1 genes, as previously reported.3,8,12 The primers used in this study are listed in Table 1.3,12 PCR was carried out in a thermocycler (Cyclogene, Techne, UK). A single reaction mixture contained: 5 µL (5 ng) of genomic DNA, 1µL (10 pmol) of each primer (Promega, USA), and 12.5 µL of PCR HotStarTaq Master Mix (Promega) containing 0.4 mM of each dNTP, 3 mM MgCl2, and 0.08 U/ųL Taq DNA polymerase in reaction buffer with a final volume of 25 µL. Initial denaturation (94°C for 4 min) was followed by 30 cycles of amplification. Each cycle consisted of 94°C for 45 seconds, various annealing temperature for different genes for 45 seconds (Table 1), and 72°C for 45 seconds. A final extension step (72°C for 5 min) completed the amplification. The amplified products were electrophoresed on 1.5% agarose gel (Sigma, USA) and were visualized by ethidium bromide staining (Sigma, USA).
 |
Table 1 Sequences of Primers Used in This Study for PCR Assays for Detection of Biofilm-Related Genes in A. baumannii Isolates (n = 94 Isolates) |
Statistical Analysis
Data were coded, validated and analysed using the Statistical Package for the Social Sciences (SPSS), version 22 (SPSS Inc., Chicago, IL, USA). Frequency, percentage, arithmetic mean and median were used to present the data. Numerical data were compared using the Student’s t-test, and categorical data were compared using the chi-squared test or Fisher’s exact test, where appropriate. All comparisons were two-tailed, and p values <0.05 were considered statistically significant. Binary logistic regression analysis with adjusted odds ratio (aOR) and antecedent 95% confidence intervals (CI) were used to identify potential risk factors.
Ethical Considerations
The present study followed the principles of the Helsinki Declaration and its amendments. Ethical approval was obtained from the Institutional Review Board (IRB) of National Liver Institute (NLI) (Protocol No. 201,807,358). All the participants were informed about the purpose of the study and written consent was obtained from all participants.
Results
Isolates Distribution
All 94 non-duplicate clinical isolates of A. baumannii during the study period were isolated from non-redundant patients (38 isolates from females and 56 isolates from males). The age of patients ranged from 41 to 71 years with an average of 55.5±12.3 and a median of 53.6 years. The specimens from which organisms were isolated included respiratory specimens (50%; 42.6% from sputum and 7.4% from endotracheal aspirate samples), blood (33%), and wound swab (17%). The majority of clinical isolates were collected from ICU (67%) followed by medical ward (16%), burn unit (10.6%), and surgical ward (6.4%).
Characteristics of Biofilm Producers
Among all A. baumannii isolates tested for biofilm formation, 66 (70.1%) isolates were biofilm producers and 28 (29.9%) isolates were non-biofilm producers. Of 66 biofilm-producing strains, 16% isolates were considered as weak biofilm producers, 34% as moderate biofilm producers and 20.2% as strong biofilm producers.
The distribution of different biofilm-producing A. baumannii phenotypes among clinical specimen types and hospital wards is presented in Figure 1. Interestingly, 66.7% (44 isolates) of biofilm producers and 53.6% (15 isolates) of non-biofilm producers were isolated from ICU. Besides, more than half of biofilm producers (23 isolates; 52%) and non-biofilm producers (16 isolates; 57%) were isolated from blood and sputum specimens, respectively.
 |
Figure 1 The distribution of different biofilm-producing A. baumannii phenotypes among (A) Clinical specimen’s types and (B) Hospital department’s types. |
All A. baumannii were investigated for the presence of biofilm-related genes (bap, ompA and blaPER-1 genes). The PCR assays yielded positive results for one gene in 41 (43.7%) isolates, and two genes in 18 (19.1%) isolates. A. baumannii isolates harbouring ompA gene with the rate of 63.8% (60 isolates) were the most prevalent genotype, while the prevalence of bap and blaPER-1 genes among isolates was 13.8% (13 isolates) and 10.6% (10 isolates), respectively.
The demographic and clinical characteristics of patients infected by biofilm-forming and non-biofilm-forming A. baumannii isolates are presented in Table 2. There were no significant differences in the demographic characteristics, types of specimen, ward admission, comorbidities, or invasive procedures between the two groups. However, the length of hospital stay, the previous antibiotic therapy, and the presence of ompA or bap genes were statistically significant risk factors associated with infections by biofilm-producing isolates (p values: 0.04, 0.015, 0.002, 0.012, respectively).
 |
Table 2 Demographic and Clinical Characteristics of Study Patients Infected by Biofilm- and Non-Biofilm-Forming A. baumannii Nosocomial Isolates |
Biofilm Production and Antibiotic Resistance
To assess whether biofilm formation is associated with any particular antimicrobial resistance pattern, all isolates of different biofilm-producing phenotypes with various resistance profiles for 16 antibiotics were compared (Table 3). The biofilm-producing isolates showed a statistically significant higher resistance rate to ceftazidime, ampicillin/sulbactam, piperacillin/tazobactam, piperacillin, gentamycin, trimethoprim/sulfamethoxazole, tigecycline and imipenem (p = 0.041, <0.001, 0.006, 0.034, 0.028, 0.002, 0.002, and 0.02, respectively). Interestingly, the presence of MDR (p = 0.017) and XDR (p = 0.002) was significantly associated with biofilm-producing capability of the isolates, compared to non-biofilm producing capabilities.
 |
Table 3 Comparison of Antimicrobial Resistance Patterns of Biofilm-Forming and Non-Biofilm-Forming A. baumannii Nosocomial Isolates |
Potential Factors Associated with Biofilm-Producing Capability
The multiple logistic regression analysis with adjusted odds ratio (aOR) and antecedent 95% confidence intervals (CI) identified potential risk factors linked to biofilm producing ability among all isolates (Table 4). The length of hospital stay (aOR= 0.023, 95% CI: 0.003–0.043), the presence of ompA gene (aOR = 0.286, 95% CI: 0.115–0.456) or bap gene (aOR = 0.346, 95% CI: 0.081–0.610), ampicillin/sulbactam resistance (aOR = 1, 95% CI: 0.652–1.348), and the presence of MDR (aOR: −0.329, 95% CI: −0.457 – −0.112) or XDR (aOR = −0.252, 95% CI: −0.328 – −0.047) were considered significant risk factors associated with biofilm producing isolates.
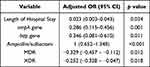 |
Table 4 Multivariate Analysis of Factors Associated with Biofilm-Forming A. baumannii Infections |
Discussion
The emerging trend of biofilm formation among MDR and XDR A. baumannii clinical isolates and its association with serious nosocomial infections has been considered a global health crisis.1,6,15 In this study, the observed overall rate of biofilm-producing A. baumannii was 70% and half of them were moderate to strong biofilm producers. These results are consistent with previous findings from other studies, which found values ranging from 50% to 76%, worldwide.2–6 Previous epidemiological studies have showed that biofilm formation is the most important virulence factor facilitating the chronicity and endemicity of A. baumannii strains in both nosocomial infections and hospital environment.3,5,12,14,22,23 It is noteworthy that >70% of biofilm-producing A. baumannii isolates in this study were obtained from hospitalized patients in ICU wards. This finding would provide an additional explanation for the role of biofilm-producing A. baumannii in nosocomial infections and outbreaks among high-risk ICU patients.3,5
Several epidemiological and molecular studies investigated the various A. baumannii repertoires of biofilm-related virulence genes and proteins which contribute to the microbial ability to adhere and form biofilms on diverse surfaces and environments.8–16 Previous reports have demonstrated that bap and ompA genes are the most prevalent genotypes in clinical and environmental isolates. In this study, about two thirds of isolates harboured ompA gene, while only 14% of isolates were positive for bap gene. These results are consistent with previous findings from other studies. Zeighami et al3 investigated a battery of biofilm-related genes in 100 MDR A. baumannii nosocomial isolates from Iranian ICU patients, and found that 81% of isolates were positive for ompA gene. Reports from other countries revealed a prevalence rate of 84.4% in Thailand,12 76.5% in Korea13 and 68.8% in Taiwan.14 These findings highlight the crucial role of ompA gene in the attachment of this pathogen to human epithelial cells and development of biofilms. Besides, other studies reported the association of ompA gene harbouring and antimicrobial resistance to individual antibiotics such as cefotaxime, aztreonam, ciprofloxacin, and imipenem via specific efflux pumps.12,14,16
Due to the continuous exposure to significant selective pressure in the hospital environment, A. baumannii usually develop acquired resistance to several antibiotic classes and subclasses through a wide variety of mechanisms, including efflux pumps, porin expression, antibiotic target mutations, and drug-inactivating enzymes.24 In this study, similar to previous reports, ≥70% of isolates were resistant to cephalosporins, aminoglycosides, carbapenems, and fluoroquinolones, which are commonly used in clinical practice. Besides, the prevalence rate of MDR and XDR isolates in this study was 28% and 73%, respectively. Worldwide, the high prevalence rates of MDR clinical isolates have been reported, ranging from 21% to 95%.3,12–15,23 Although determining the prevalence of XDR A. baumannii is challenging, and not fully addressed, carbapenem resistance has been considered one of the hallmarks of XDR A. baumannii.25 According to CDC/NHSN, the overall rate of carbapenem-resistant isolates is 47–64% in USA.26 Reports from other parts of the world showed a prevalence rate of 49% in Europe,27 40–60% in Southeast Asia28 and 40–80% in Latin America.29 The overall resistance rate to tigecycline and colistin in this study is higher than that reported in previous studies.15,25 This finding is worrisome and of great concern as these two antibiotics have been considered as the last resort drugs for the treatment of A. baumannii nosocomial infections in either single or combined chemotherapeutic regimens.7,25
It is worth noting that the resistance rate to individual antibiotics as well as the prevalence rate of MDR and XDR in biofilm-producing isolates in this study was higher than that in non-biofilm producers. Previous studies have reported a positive relationship between biofilm formation and antibiotic resistance in A. baumannii isolates with higher resistance rates to aminoglycosides, carbapenems, and sulphonamides. For example, Durate et al25 found that the isolates resistant to gentamicin and tobramycin were more frequently able to form biofilms (74% and 73.3%, respectively). Thummeepak et al12 found that 125 (81.7%) of 153 gentamicin resistant clinical isolates were biofilm producers. A study performed on clinical isolates from patients with nosocomial infections in three Iranian hospitals found that 92% of 155 biofilm-forming isolates were MDR.30 Moreover, Zeighami et al3 reported that 49 of 100 biofilm-producing A. baumannii clinical isolates were strong biofilm producers and XDR.
To better understand the pathogenic and epidemiologic role of biofilm-producing isolates, regression analysis was used to identify the potential risk factors linked to biofilm-producing ability among all isolates. The risk factors associated significantly with biofilm production of nosocomial isolates in this study were the length of hospital stay (P = 0.03), the presence of ompA gene (P = 0.001) or bap gene (P = 0.01), ampicillin/sulbactam resistance (P = <0.001), and the presence of MDR (P = 0.01) or XDR (P = 0.02). Although part of these findings have already been described in previous studies as risk factors linked to hospital-acquired infection by various nosocomial pathogens,4,5,26,31 our results could be considered as useful predictors for the acquisition of nosocomial infections by biofilm-producing A. baumannii clinical isolates. These findings would help clinicians design necessary actions for prudent antibiotic policies and effective infection control measure inside hospitals.
This study had some limitations. Firstly, environmental isolates were not included and biofilm-related genes other than ompA, bap and blaPER-1 genes were not investigated. However, this study could be considered as the first epidemiological report including the most prevalent biofilm genetic determinants among nosocomial isolates in our region. Secondly, the lack of molecular genotyping and genome sequencing in this study may have limited its ability to deeply explore the pathogenic role of other virulence markers genes. Finally, this was a single centre study. Therefore, our findings may not be generalized to other settings. Further molecular-based epidemiological multi-centre studies of longer surveillance duration are necessary to better understand the phenotypic and genotypic correlations between biofilm-related virulence genes and antimicrobial resistance. These studies are necessary to help determine national priorities for local interventions.
Conclusion
The high prevalence of biofilm-producing MDR and XDR nosocomial isolates in this study is worrisome and alarming. The high resistance rate to tigecycline and colistin is challenging and of great concern. Our results highlighted the importance of the length of hospital stay, the presence of ompA or bap genes, ampicillin/sulbactam resistance, and the presence of MDR or XDR as potential risk factors associated with biofilm-producing A. baumannii clinical isolates. These findings could be beneficial for health authorities and decision makers to better control the continuous selection and transfer of this serious phenotype inside hospitals and to improve the quality of patients’ care.
Funding
The authors declare that no financial support was obtained from any funding organizations or pharmaceutical companies.
Disclosure
The authors declare no conflicts of interests for this work.
References
1. Wong D, Nielsen TB, Bonomo RA, et al. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev. 2017;30:409–430.
2. Bahador A, Farshadzadeh Z, Raoofian R, et al. Association of virulence gene expression with colistin-resistance in Acinetobacter baumannii: analysis of genotype, antimicrobial susceptibility, and biofilm formation. Ann Clin Microbiol Antimicrob. 2018;17(1):24–35. doi:10.1186/s12941-018-0277-6
3. Zeighami H, Valadkhani F, Shapouri R, et al. Virulence characteristics of multidrug resistant biofilm forming Acinetobacter baumannii isolated from intensive care unit patients. BMC Infect Dis. 2019;19(1):629–637. doi:10.1186/s12879-019-4272-0
4. Sunenshine RH, Wright MO, Maragakis LL, et al. Multidrug-resistant Acinetobacter infection mortality rate and length of hospitalization. Emerg Infect Dis. 2007;13(1):97–103. doi:10.3201/eid1301.060716
5. Weber DJ, Rutala WA, Miller MB, et al. Role of hospital surfaces in the transmission of emerging health care-associated pathogens: norovirus, Clostridium difficile, and Acinetobacter species. Am J Infect Control. 2010;38(Suppl 5):S25–33. doi:10.1016/j.ajic.2010.04.196
6. World Health Organization. Media centre. WHO publishes list of bacteria for which new antibiotics are urgently needed; 2017. Available from: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/.
7. Lee CR, Lee JH, Park M, et al. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol. 2017;7:55.
8. Bardbari AM, Arabestani MR, Karami M, et al. Correlation between ability of biofilm formation with their responsible genes and MDR patterns in clinical and environmental Acinetobacter baumannii isolates. Microb Pathog. 2017;108:122–128. doi:10.1016/j.micpath.2017.04.039
9. Vila-Farres Z, Parra-Millan R, Sanchez-Encinales V, et al. Combating virulence of gram-negative bacilli by ompA inhibition. Sci Rep. 2017;7(1):14683. doi:10.1038/s41598-017-14972-y
10. Greene C, Vadlamudi G, Newton D, et al. The influence of biofilm formation and multidrug resistance on environmental survival of clinical and environmental isolates of Acinetobacter baumannii. Am J Infect Control. 2016;44(5):e65–71. doi:10.1016/j.ajic.2015.12.012
11. Jahangiria S, Malekzadegana Y, Motamedifara M, et al. Virulence genes profile and biofilm formation ability of Acinetobacter baumannii strains isolated from inpatients of a tertiary care hospital in southwest of Iran. Gene Rep. 2019;17:100481. doi:10.1016/j.genrep.2019.100481
12. Thummeepak R, Kongthai P, Leungtongkam U, et al. Distribution of virulence genes involved in biofilm formation in multi-drug resistant Acinetobacter baumannii clinical isolates. Int Microbiol. 2016;19(2):121–129. doi:10.2436/20.1501.01.270
13. Yang CH, Su PW, Moi S, et al. Biofilm formation in Acinetobacter baumannii: genotype-phenotype correlation. Molecules. 2019;24(10):E1849. doi:10.3390/molecules24101849
14. Sung JY. Molecular characterization and antimicrobial susceptibility of biofilm-forming Acinetobacter baumannii clinical isolates from Daejeon, Korea. Korean J Clin Lab Sci. 2018;50(2):100–109. doi:10.15324/kjcls.2018.50.2.100
15. Salehi B, Goudarzi H, Nikmanesh B, et al. Emergence and characterization of nosocomial multidrug-resistant and extensively drug-resistant Acinetobacter baumannii isolates in Tehran, Iran. J Infect Chemother. 2018;24(7):515–523. doi:10.1016/j.jiac.2018.02.009
16. Smani Y, Fabrega A, Roca I, et al. Role of ompA in the multidrug resistance phenotype of Acinetobacter baumannii. Antimicrob Agents Chemother. 2014;58(3):1806–1808. doi:10.1128/AAC.02101-13
17. STROBE checklist, version 4; 2007. Available from: http://www.strobe-statement.org/index.php?id=available-checklists.
18. Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008;36(5):309–332. doi:10.1016/j.ajic.2008.03.002
19. Carrol KC, Pfaller MA, Landry ML, et al. Manual of Clinical Microbiology Handbook.
20. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing.
21. Pfizer Inc. (Wyeth Pharmaceuticals). Tygacil® product insert. Philadelphia, PA; 2016. Available from: http://labeling.pfizer.com/showlabeling.aspx?id=491&pagename=tygacil_fly.
22. Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug resistant and pan drug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
23. Monfared AM, Rezaei A, Poursina F, et al. Detection of genes involved in biofilm formation in MDR and XDR Acinetobacter baumannii isolated from human clinical specimens in Isfahan, Iran. Arch Clin Infect Dis. 2019;14(2):e85766. doi:10.5812/archcid.96531
24. Eichenberger EM, Thaden JT. Epidemiology and mechanisms of resistance of extensively resistant gram-negative bacteria. Antibiotics (Basel). 2019;8(2):E37. doi:10.3390/antibiotics8020037
25. Duartea A, Ferreiraa S, Almeidaa S, et al. Clinical isolates of Acinetobacter baumannii from a Portuguese hospital: PFGE characterization, antibiotic susceptibility and biofilm-forming ability. Comp Immunol Microbiol Infect Dis. 2016;45:29–33. doi:10.1016/j.cimid.2016.02.002
26. Weiner LM, Webb AK, Limbago B, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2011–2014. Infect Control Hosp Epidemiol. 2016;37(11):1288–1301. doi:10.1017/ice.2016.174
27. European Centre for Disease Prevention and Control. Surveillance of antimicrobial resistance in Europe 2017. Sona, Sweden: European Center for Disease Prevention. Available from: https://ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2017.
28. Hsu LY, Apisarnthanarak A, Khan E, et al. Carbapenem-resistant Acinetobacter baumannii and enterobacteriaceae in south and southeast Asia. Clin Microbiol Rev. 2017;30(1):1–22. doi:10.1128/CMR.00042-16
29. Rodriguez CH, Nastro M, Famiglietti A. Carbapenemases in Acinetobacter baumannii. Review of their dissemination in Latin America. Rev Argent Microbiol. 2018;50(3):327–333. doi:10.1016/j.ram.2017.10.006
30. Babapour E, Haddadi A, Mirnejad R, et al. Biofilm formation in clinical isolates of nosocomial Acinetobacter baumannii and its relationship with multidrug resistance. Asian Pac J Trop Biomed. 2016;6(6):528–533. doi:10.1016/j.apjtb.2016.04.006
31. Wang L, Zhou K, Chen W, et al. Epidemiology and risk factors for nosocomial infection in the respiratory intensive care unit of a teaching hospital in China: a prospective surveillance during 2013 and 2015. BMC Infect Dis. 2019;19(1):145–153. doi:10.1186/s12879-019-3772-2
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
