Back to Journals » Infection and Drug Resistance » Volume 16
Epidemiological and Molecular Characteristics of blaNDM-1 and blaKPC-2 Co-Occurrence Carbapenem-Resistant Klebsiella pneumoniae
Authors Rong F, Liu Z, Yang P, Wu F, Sun Y, Sun X, Zhou J
Received 3 December 2022
Accepted for publication 1 April 2023
Published 17 April 2023 Volume 2023:16 Pages 2247—2258
DOI https://doi.org/10.2147/IDR.S400138
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Fang Rong,1,2,* Ziyi Liu,3,4,* Pengbin Yang,3,4 Feng Wu,5 Yu Sun,5 Xuewei Sun,5 Jun Zhou5
1Department of General Practice, The Affiliated Hospital of Yangzhou University, Yangzhou, Jiangsu, People’s Republic of China; 2Graduate School Department of Dalian Medical University, Dalian, Liaoning, People’s Republic of China; 3Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, College of Veterinary Medicine, Yangzhou University, Yangzhou, Jiangsu, People’s Republic of China; 4Institute of Comparative Medicine, Yangzhou University, Yangzhou, Jiangsu, People’s Republic of China; 5Department of Respiratory and Critical Care Medicine, The Affiliated Hospital of Yangzhou University, Yangzhou University, Yangzhou, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jun Zhou, Department of Respiratory and Critical Care Medicine, The Affiliated Hospital of Yangzhou University, Yangzhou University, 368 Hanjiang Middle Road, Yangzhou, Jiangsu, 225009, People’s Republic of China, Email [email protected]
Objective: Carbapenem-resistant Klebsiella pneumoniae (CRKP) has emerged and spread worldwide. It can usually cause a serious threat complicating treatment options in clinical settings. However, treatment options are limited. The present study investigates the prevalence and genetic characteristics of blaNDM-1 and blaKPC-2 co-harboring clinical isolates of Klebsiella pneumoniae.
Methods: In this study, Multiplex polymerase chain reaction (PCR) was performed to detect the carbapenem-resistant genes, and the broth microdilution method was used to determine the minimum inhibitory concentrations (MICs) of antibacterial drugs. The transferability of carbapenem-resistant phenotypes was examined using filter mating assays. Overall, we used Illumina sequencing to evaluate the epidemiological and molecular characteristics of blaNDM-1 and blaKPC-2 (genes encoding carbapenemase) co-occurrence in CRKP strains.
Results: All strains exhibited resistance to carbapenems and other antibiotics. However, they were still susceptible to polymyxin E. Among them, 18 isolates were positive for blaKPC-2, blaNDM-1, and multiple virulence determinants, such as genes encoding the virulence factor aerobactin, yersiniabactin, and the regulator of the mucoid phenotype (rmpA and rmpA2). Whole genome sequencing revealed that the 18 CRKP strains belonged to ST11 and capsular serotype KL64, and could be grouped into two evolutionary branches. Furthermore, these strains displayed hypervirulence potential since all of them carried pLVPK-like plasmid.
Conclusion: These findings suggested that ST11-KL64 CRKP strains are major threats in terms of nosocomial infections in this hospital. Hence, new strategies should be urgently developed to monitor, diagnose, and treat this high-risk CRKP clone.
Keywords: Klebsiella pneumoniae, carbapenem resistance, illumina sequencing, blaKPC-2, blaNDM-1
Introduction
Klebsiella pneumoniae is an important clinical pathogen. With the widespread use of antibiotics, its multidrug resistance has gradually increased, particularly to carbapenems.1 The first carbapenemase in K. pneumoniae was discovered in 1996, which was encoded by blaKPC gene.2 Subsequently, other carbapenemase genes were discovered, including blaNDM, blaOXA-48, blaVIM, and blaIMP.3–6 Among them, KPC-2, one of class A carbapenemase, is relatively common in China. With the widespread popularity of carbapenem and wide distribution of carbapenemase genes, carbapenem-resistant K. pneumoniae (CRKP) isolation rates are gradually increasing globally. At present, K. pneumoniae carbapenemase (KPC) is one of the most important carbapenemases in clinical setting, and its rapid spread has posed a huge challenge to global public health.7 In China, CRKP isolates mainly carry blaKPC-2 or blaNDM-1 genes for carbapenem resistance, and multilocus sequence typing is mainly ST11.8,9
K. pneumoniae exhibits two pathogenic types: hypervirulent K. pneumoniae (hvKP) and classical K. pneumoniae (cKP).10 These two types cause a great challenge globally in terms of hospital infections.11 cKP forms multidrug-resistant (MDR) and extensively drug-resistant strains by acquiring various antimicrobial resistance genes.1 A typical representative is CRKP, which is related with high morbidity and mortality rates.12 hvHP easily causes liver abscess, pneumonia, meningitis, and endophthalmitis in healthy individuals and has been reported to be closely related to iconic virulence factors including regulators of myxoid phenotypes (rmpA and rmpA2) and virulence genes (yersiniabactin and aerobactin).10 For a long time, K. pneumoniae did not encode both multidrug resistance and high virulence phenotypes simultaneously.13 However, in recent years, reports are emerging continuously regarding the simultaneous emergence of carbapenem resistance and virulence in a single epidemic clone, which has become a serious public health threat.14–16 ST11-CR-hvKp is the most representative clade detected in China.15,17,18 Therefore, the genomic characterization of CRKP should be urgently studied to prevent, diagnose, and treat infections caused by K. pneumoniae.
In this study, we obtained 60 CRKP isolates from inpatients in a tertiary hospital in China, among which the NDM and KPC co-producing ST11-KL64 CRKP clone is a major threat in terms of nosocomial infections at this hospital, and these
strains displayed hypervirulence potential since all of them carried pLVPK-like plasmid. In addition, the transmission routes and genetic characterizations of these strains were investigated as well.
Materials and Methods
Patients and Isolates
From January 2019 to June 2021, 60 CRKP isolates were isolated from sputum, Tracheal aspiration, BAL, blood, urine, pus, and ascites fluid samples from the general ward and intensive care unit (ICU) of a tertiary hospital in Yangzhou, Jiangsu Province, China. There are 35 beds in the ICU of the hospital. The clinical data of patients were reviewed from the patient management system, including gender, age, in-patient department, underlying disease, clinical diagnosis, prognosis, duration of hospital stay, invasive procedures performed before sample collection, and antibiotic exposure. The underlying diseases included diseases of immune, respiratory, urinary, digestive, and blood systems, as well as hypertension, diabetes, and tumors. Invasive operations included invasive ventilator, tracheotomy and intubation, and drainage. The study was approved by the Research Ethics Committee of the Affiliated Hospital of Yangzhou University and was followed the Declaration of Helsinki. No identifiable patient information was collected in this study. All isolates were isolated for routine clinical experiment.
Bacterial Identification and Antimicrobial Susceptibility
The samples were plated onto 5% sheep blood agar and were cultured at 37°C for bacterial isolation. All isolates were identified using the VITEK-2 system. A multiplex PCR method was used to assess whether these isolates contained carbapenemase genes including blaNDM, blaKPC, blaSIM, blaGIM, blaSPM, blaAIM, blaOXA-48, blaBIC, blaDIM, blaIMP, blaVIM (Table S1), referring to the primer sequence.19 The isolates carrying the corresponding carbapenemase-encoding gene were used as positive controls. Antimicrobial susceptibility was investigated using VITEK-2 system and the broth microdilution methods. The susceptibility breakpoint was interpreted according to 2018 Clinical and Laboratory Standards Institute guideline except for tigecycline and polymyxin E, which followed the criteria of European Committee on Antimicrobial Susceptibility Testing (version 12.0). Escherichia coli ATCC25922 was used as the quality control strain for antimicrobial susceptibility testing.
Filter Mating Assay
The transferability of carbapenem-resistant phenotypes was measured using filter-mating assay. K. pneumoniae YZ6 Hygr was used as the recipient strain, and the 60 CRKP isolates were used as the donor strains. Transconjugants were selected on LB agar plates supplemented with hygromycin (200 mg/L) and meropenem (2 mg/L).20 The transconjugants carrying the gene encoding carbapenemase were confirmed using multiplex PCR and antimicrobial susceptibility testing.
Genome Extraction, Bioinformatics Analysis, and Phylogenetic Tree Construction
The epidemiological and molecular features of blaKPC and blaNDM co-occurrence were further studied using 18 CRKP isolates with blaKPC-2 and blaNDM-1 co-occurrence. FastPure Bacteria DNA Isolation Mini Kit (Vazyme, Nanjing, China) was used to extract the genomic DNA from all CRKP isolates with blaKPC-2 and blaNDM-1 co-occurrence. The extracted genomic DNA was subjected to 1% agarose gel electrophoresis, quantified using the Qubit fluorometer, and further subjected to short-read sequencing (2 × 150 bp) on the Illumina HiSeq 2500 platform. The short-read Illumina raw sequences of 18 CRKP isolates with blaKPC-2 and blaNDM-1 co-occurrence were screened for quality and assembled through SPAdes. The contigs smaller than 500 bp were purposefully removed. The sequence types (STs), capsule types, insertion sequences (IS), and multiple antimicrobial-resistance and virulence genes of these isolates were identified using CGE server (https://cge.cbs.dtu.dk) and Kleborate. The phylogenetic trees for the 18 CRKP isolates with blaKPC-2 and blaNDM-1 co-occurrence were constructed using Roary and FastTree. Further, visualization and modification were performed using iTOL5.
Results
Clinical Characteristics of CRKP Isolates
From January 2019 to June 2021, 60 strains of CRKP were isolated from 54 inpatients in a tertiary hospital in Yangzhou (Table 1). Among the 54 inpatients, 39 were males (72.22%) and 15 were females (27.78%), with an average age of 65.0 years. Clinical isolates were obtained from sputum (35/60, 58.33%), Tracheal aspiration (8/60,13,33%), BAL (5/60,8.34%), blood (6/60, 10.00%), urine (4/60, 6.67%), pus (1/60, 1.67%), and ascites (1/60, 1.67%) samples. The inpatients were from ICU (38/54, 70.37%), respiratory medicine (3/54, 5.56%), neurosurgery (3/54, 5.56%), general surgery (3/54, 5.56%), emergency (2/54, 3.70%), thoracic surgery (1/54, 1.85%), neurology (1/54, 1.85%), gastroenterology (1/54, 1.85%), cardiology (1/54, 1.85%), and nephrology (1/54, 1.85%) departments. Most patients were exposed to antibiotics, mainly cephalosporins, carbapenems, and enzyme inhibitors, and 45 patients had undergone invasive operation before sample collection.
 |
Table 1 Characteristics of the 60 CRKP Isolates from the 54 Inpatients |
Antimicrobial Susceptibility and Transferability
According to the drug sensitivity test data (Table 2), all CRKP isolates exhibited multiple-drug resistance. Moreover, most strains displayed resistance to doxycycline (58/60, 96.7%), chloramphenicol (59/60, 98.3%), and aztreonam (59/60, 98.3%) but remained susceptible to polymyxin (59/60, 98.3%). Multiple PCR revealed that a total of 18 isolates simultaneously carried blaNDM-1 and blaKPC-2. The results of conjugation assay (Figure 1) revealed that most of the blaKPC-2-carrying CRKP isolates could not successfully transfer their carbapenemase genes into the recipient strain. By contrast, among the 18 strains with blaKPC-2 and blaNDM-1 co-occurrence, 16 strains could successfully transfer the carbapenem resistance phenotype to the recipient strain YZ6 Hygr, indicating that their carbapenem encoding genes were located on the conjugate plasmids.
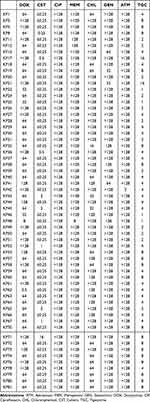 |
Table 2 MICs (Mg/L) of All CRKP Strains in This Study |
STs, Capsular Types, Virulence Genes, and Phylogenetic Analysis of CRKP Strains with blaNDM-1 and blaKPC-2 Co-Occurrence
MLST is a well-known gene typing method that can be used to monitor and control the spread of pathogens in hospitals.21 According to the K. pneumoniae MLST database, 18 CRKP strains with blaNDM-1 and blaKPC-2 co-occurrence were all identified as ST11, and the capsular type were KL64, suggesting that there was nosocomial infection of ST11 CRKP at this hospital. In addition, phylogenetic analysis revealed that 18 CRKP isolates with blaNDM-1 and blaKPC-2 co-occurrence were divided into two clades (Figure 2). Clade 1 included 9 strains (KP72, KP74, KP37, KP60, KP52, KP63, KP53, KP64 and KP62), whereas clade 2 contained 9 strains (KP58, KP44, KP43, KP80, KP78, KP20, KP81, KP73, and KP57).
Identification of Antimicrobial Resistance Genes and Virulence Genes
According to the WGS results, the antibiotic-resistance genes and virulence genes of 18 CRKP strains with blaNDM-1 and blaKPC-2 co-occurrence were identified (Figure 2). All isolates contained resistance genes against β-lactams (blaCTX-M-65, blaKPC-2), tetracyclines [tet(A)], aminoglycosides (rmtB), sulfonamides (sul2), trimethoprim (dfrA14), fosfomycin (fosA), and quinolones (qnrS1). The β-lactam resistance gene blaTEM-1B was present in 17 CRKP isolates; however, it was not present in KP74 isolates. The resistance genes against β-lactams (blaADC-25, blaOXA-23, and blaOXA-66), aminoglycosides [aph(3′)-Ic, strB, and strA], tetracyclines [tet(B)], macrolides [mph(E), msr(E)] were observed in KP62 isolates. However, these resistance genes were not observed in the remaining 17 isolates. Analysis combined with clinical data (Table 1), KP62 strains were isolated from ICU, and the patients had undergone invasive operations before sample collection. In addition, all CRKP strains with blaNDM-1 and blaKPC-2 co-occurrence contained yersiniabactin and aerobactin, which represented the potential of CR-hvKP phenotype. Except KP62, the regulatory factors of mucus phenotype gene (rmpA2) were detected in all CRKP isolates with blaNDM-1 and blaKPC-2 co-occurrence.
To determine the type of virulence plasmid harbored by these strains, plasmid pLVPK (GenBank accession AY378100), a classical virulence plasmid carrying a set of virulence genes, including iroBCDN, iucABCD, rmpA, and rmpA2, was used as reference plasmid. Surprisingly, we found that the virulence plasmids carried by all strains were aligned well with pLVPK based on Illumina-based contigs analysis (Figure 3), suggesting the identified virulence genes in these strains might be closely related to pLVPK-like plasmids.
Discussion
ICU is the main site of nosocomial infections and has been generally considered as a suitable place to study the epidemic characteristics of MDR strains,22,23 particularly K. pneumoniae.24 The phenomenon of drug resistance in bacterial isolates from ICU is becoming more and more serious in recent years. It is closely related to factors such as inpatients suffering from various basic diseases, the use of invasive surgical treatment, prolonged hospitalization, and extensive use of broad-spectrum antibiotics, causing a great challenge in terms of clinical antibacterial treatment.25,26 In the study, among the 60 CRKP strains, 42 (70.00%) were isolated from the ICU. This exhibited a high prevalence rate in inpatients, which was consistent with previous studies.27,28
According to MLST typing, all 18 CRKP strains with blaNDM-1 and blaKPC-2 co-occurrence belonged to ST11, indicating that K. pneumoniae positive for ST11 blaKPC-2 was the dominant strain in this hospital. This was consistent with previous studies in China.27,29 ST11 is the single locus variant of ST258, and they belong to the same clonal members of CG258,7 which has greatly contributed to the global spread of KPC-producing CRKP during the past 20 years.9,30 In contrast, the ST258 is prevalent in North America and Europe, whereas ST11 is the main type in Asia.31,32 Additionally, ST11 is the main sequence type of KPC producing CRKP in China and has been reported globally, including the United States, Europe, and Asia.9,12,31–37
Previously, K. pneumoniae with the ST11 phenotype was a widely and commonly occurring MDR clone, exhibiting resistance to carbapenems; however, it was not highly virulent. However, ST11 has recently attracted considerable attention because of its feature of co-occurrence of resistance and hypervirulence genes in a single strain.12 In our study, the analysis by Kleborate revealed that all 18 CRKP strains with blaNDM-1 and blaKPC-2 co-occurrence exhibited synthesis of aerobactin, which has been considered as the major siderophore system in the hvKP. The rmpA and rmpA2 virulence genes have been thought to control the capsular polysaccharide biosynthesis and symbolize a hypermucoviscous phenotype, which also existed in most of the 18 strains. Therefore, 18 CRKP strains with blaNDM-1 and blaKPC-2 co-occurrence isolated in this study exhibited hypervirulence phenotype and deserved our attention.
In addition, CRKP strains with blaNDM-1 and blaKPC-2 co-occurrence belonged to serotype KL64 in this study, which was different from previously reported KL1, KL2, and KL62 serotypes.9 Clinically, the ST11-KL64 CRKP isolates carrying the rmpA and rmpA2 virulence genes and producing KPC-2 were more survivable in the environment and could cause more severe infection.34 It is reported that the inpatients infected with ST11-KL64 CRKP had a higher mortality. The results of this study revealed that one patient died among 16 patients with the infection of ST11-KL64 CRKP carrying rmpA and rmpA2 genes, indicating a high mortality rate of ST11-KL64 CRKP. This revealed the highly virulent nature of these ST11-KL64 CRKP isolates, and targeted surveillance was urgently needed in this regard. It is necessary to further conduct genomic epidemiological and evolutionary analyses throughout the country to elucidate the genetic basis and evolutionary characteristics of the widely spread carbapenem-resistant and highly virulent ST11-KL64 K. pneumoniae in China.
The results of phylogenetic tree revealed that 18 ST11-KL64 CRKP strains with blaNDM-1 and blaKPC-2 co-occurrence could be divided into two evolutionary clades, indicating two independent transmission events. Some patients of the two evolutionary clades overlapped in terms of strain isolation time or inpatient departments, which may be the main reason for the spread of K. pneumoniae in this hospital. For example, KP43, KP53, KP57, KP60, KP62, and KP64 were isolated from 8 hospitalized ICU patients with similar sampling time, indicating that transmission events occurred in a short period of time. The spread of ST11-CRKP in different departments or different wards of the same department in the hospital has been frequently reported.6,9,11,32,38 The results revealed two independent outbreaks of ST11-KL64 CRKP strains with blaNDM-1 and blaKPC-2 co-occurrence in the ICU and respiratory ward from 2019 to 2021. These results confirmed that ST11-CRKP strains were prone to transfer. Further, we should further analyze the drug-resistant gene transfer of coexisting strains, and analyze the genetic environment of blaNDM-1 and blaKPC-2 in combination with the long read sequencing results. Therefore, practical approaches must be implemented to control transmission and reduce the occurrence of nosocomial infections.
Multiplex PCR revealed that among 60 CRKP isolates, 54 strains produced carbapenemase and contained blaKPC gene. This was the most common mechanism of carbapenem resistance in K. pneumoniae. In China, it is reported that KPC-producing K. pneumoniae was the main strain causing outbreak.39,40 Among the 54 CRKP strains with KPC-producing gene, 18 strains carried blaNDM gene at the same time. In addition, the Illumina sequencing analysis revealed that apart from the many types of β-lactam-resistance genes, other resistance genes such as tet(A), rmtB, sul2, dfrA14, fosA, and qnrS1 were present in all 18 ST11-KL64 CRKP isolates with blaNDM-1 and blaKPC-2 co-occurrence, conferring resistance to tetracyclines, aminoglycosides, sulfonamides, trimethoprim, fosfomycin, and quinolones, respectively. The co-existence of carbapenemase, β-lactamases, and many types of drug resistance genes led to the multidrug resistance. Undoubtedly, the existence of resistance genes enables K. pneumoniae isolates to survive the attack of antibacterial drugs. Thus, treatment of infections caused by these multi-resistant CRKP strains is a great challenge because of limited availability of antimicrobials. Fortunately, polymyxin was effective in vitro, suggesting that it might be a valuable therapeutic option for ST11-KL64 CRKP infections.
Conclusion
Our study confirmed that the CRKP strains isolated from the hospital mainly had ST11-KL64 phenotype and mostly carried blaKPC-2 resistance genes. The ICU was the main site of nosocomial infection and rapid transmission of CRKP. The strains exhibited high pathogenicity by acquiring various drug-resistance genes and virulence genes, causing a major challenge to public health. Therefore, it is urgent to develop effective strategies to control and prevent further nosocomial infection.
Data Sharing Statement
The datasets presented in this study can be found in online (https://doi.org/10.6084/m9.figshare.21360120).
Acknowledgments
This work was supported by the Open Project Program of Jiangsu Key Laboratory of Zoonosis (No. R2202).
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This work was funded by the Clinical Translational Research Project of the Medical Innovation and Translation Special Fund [grant numbers AHYZUZHXM,202106].
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–275. doi:10.1093/femsre/fux013
2. Yigit H, Queenan AM, Anderson GJ, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–1161. doi:10.1128/AAC.45.4.1151-1161.2001
3. Nishida S, Matsunaga N, Kamimura Y, et al. Emergence of Enterobacter cloacae complex co-producing IMP-10 and CTX-M, and Klebsiella pneumoniae producing VIM-1 in clinical isolates in Japan. Microorganisms. 2020;8(11):1816. doi:10.3390/microorganisms8111816
4. Pérez-Vázquez M, Sola Campoy PJ, Ortega A, et al; Spanish NDM Study Group. Emergence of NDM-producing Klebsiella pneumoniae and Escherichia coli in Spain: phylogeny, resistome, virulence and plasmids encoding blaNDM-like genes as determined by WGS. J Antimicrob Chemother. 2019;74(12):3489–3496. doi:10.1093/jac/dkz366
5. Fukigai S, Alba J, Kimura S, et al. Nosocomial outbreak of genetically related IMP-1 beta-lactamase-producing Klebsiella pneumoniae in a general hospital in Japan. Int J Antimicrob Agents. 2007;29(3):306–310. doi:10.1016/j.ijantimicag.2006.10.011
6. Lu MC, Chen YT, Tang HL, et al. Transmission and evolution of OXA-48-producing Klebsiella pneumoniae ST11 in a single hospital in Taiwan. J Antimicrob Chemother. 2020;75:318–326. doi:10.1093/jac/dkz431
7. Chen L, Mathema B, Chavda KD, et al. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22(12):686–696. doi:10.1016/j.tim.2014.09.003
8. Zhang P, Shi Q, Hu H, et al. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin Microbiol Infect. 2020;26(1):124.e1–124.e4. doi:10.1016/j.cmi.2019.08.020
9. Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. doi:10.1016/S1473-3099(17)30489-9
10. Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Virulence. 2013;4(2):107–118. doi:10.4161/viru.22718
11. Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3):e00001–e00019. doi:10.1128/CMR.00001-19
12. Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18:344–359. doi:10.1038/s41579-019-0315-1
13. Yang X, Dong N, Chan EW, et al. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 2021;29(1):65–83. doi:10.1016/j.tim.2020.04.012
14. Chen L, Kreiswirth BN. Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. Lancet Infect Dis. 2018;18(1):2–3. doi:10.1016/S1473-3099(17)30517-0
15. Wong MHY, Shum HP, Chen JHK, et al. Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2018;18(1):24. doi:10.1016/S1473-3099(17)30629-1
16. Xie M, Dong N, Chen K, et al. A hybrid plasmid formed by recombination of a virulence plasmid and a resistance plasmid in Klebsiella pneumoniae. J Glob Antimicrob Resist. 2020;23:466–470. doi:10.1016/j.jgar.2020.10.018
17. Yao H, Qin S, Chen S, et al. Emergence of carbapenem-resistant hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2018;18:25.
18. Xu M, Fu Y, Fang Y, et al. High prevalence of KPC-2-producing hypervirulent Klebsiella pneumoniae causing meningitis in Eastern China. Infect Drug Resist. 2019;12:641–653. doi:10.2147/IDR.S191892
19. Poirel L, Walsh TR, Cuvillier V, et al. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi:10.1016/j.diagmicrobio.2010.12.002
20. Chen R, Liu Z, Xu P, et al. Deciphering the epidemiological characteristics and molecular features of blaKPC-2- or blaNDM-1-positive Klebsiella pneumoniae isolates in a newly established hospital. Front Microbiol. 2021;12:741093. doi:10.3389/fmicb.2021.741093
21. Baraniak A, Grabowska A, Izdebski R, et al.; KPC-PL Study Group. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008–2009. Antimicrob Agents Chemother. 2011;55(12):5493–5499. doi:10.1128/AAC.05118-11
22. Hu Y, Liu C, Shen Z, et al. Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008–2018. Emerg Microbes Infect. 2020;9(1):1771–1779. doi:10.1080/22221751.2020.1799721
23. Wang R, Yang Q, Zhang S, et al. Trends and correlation of antibiotic susceptibility and antibiotic consumption at a large teaching hospital in China (2007–2016): a surveillance study. Ther Clin Risk Manag. 2019;15:1019–1027. doi:10.2147/TCRM.S210872
24. Singh A, Goering RV, Simjee S, et al. Application of molecular techniques to the study of hospital infection. Clin Microbiol Rev. 2006;19(3):512–530. doi:10.1128/CMR.00025-05
25. Gopalakrishnan S, Kamalanathan A, Rajan S, et al. Emergence of armA and rmtB genes among VIM, NDM, and IMP metallo-β-lactamase-producing multidrug-resistant gram-negative pathogens. Acta Microbiol Immunol Hung. 2018;65(1):107–118. doi:10.1556/030.64.2017.027
26. Zhu WM, Yuan Z, Zhou HY. Risk factors for carbapenem-resistant Klebsiella pneumoniae infection relative to two types of control patients: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2020;9:23. doi:10.1186/s13756-020-0686-0
27. Qin X, Wu S, Hao M, et al. The colonization of carbapenem-resistant Klebsiella pneumoniae: epidemiology, resistance mechanisms, and risk factors in patients admitted to intensive care units in China. J Infect Dis. 2020;221(Suppl 2):S206–S214. doi:10.1093/infdis/jiz622
28. Yu F, Hu L, Zhong Q, et al. Dissemination of Klebsiella pneumoniae ST11 isolates with carbapenem resistance in integrated and emergency intensive care units in a Chinese tertiary hospital. J Med Microbiol. 2019;68(6):882–889. doi:10.1099/jmm.0.000981
29. Yang Q, Jia X, Zhou M, et al. Emergence of ST11-K47 and ST11-K64 hypervirulent carbapenem-resistant Klebsiella pneumoniae in bacterial liver abscesses from China: a molecular, biological, and epidemiological study. Emerg Microbes Infect. 2020;9(1):320–331. doi:10.1080/22221751.2020.1721334
30. Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796. doi:10.1016/S1473-3099(13)70190-7
31. Ko KS. Antibiotic-resistant clones in gram-negative pathogens: presence of global clones in Korea. J Microbiol. 2019;57(3):195–202. doi:10.1007/s12275-019-8491-2
32. Zhan L, Wang S, Guo Y, et al. Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a tertiary hospital in China. Front Cell Infect Microbiol. 2017;7:182. doi:10.3389/fcimb.2017.00182
33. Yang Y, Yang Y, Chen G, et al. Molecular characterization of carbapenem-resistant and virulent plasmids in Klebsiella pneumoniae from patients with bloodstream infections in China. Emerg Microbes Infect. 2021;10(1):700–709. doi:10.1080/22221751.2021.1906163
34. Zhou K, Xiao T, David S, et al. Novel subclone of carbapenem-resistant Klebsiella pneumoniae sequence type 11 with enhanced virulence and transmissibility, China. Emerg Infect Dis. 2020;26(2):289–297. doi:10.3201/eid2602.190594
35. Jin X, Chen Q, Shen F, et al. Resistance evolution of hypervirulent carbapenem-resistant Klebsiella pneumoniae ST11 during treatment with tigecycline and polymyxin. Emerg Microbes Infect. 2021;10(1):1129–1136. doi:10.1080/22221751.2021.1937327
36. Jiang Y, Wei Z, Wang Y, et al. Tracking a hospital outbreak of KPC-producing ST11 Klebsiella pneumoniae with whole genome sequencing. Clin Microbiol Infect. 2015;21(11):882–889. doi:10.1016/j.cmi.2015.07.001
37. Spencer MD, Winglee K, Passaretti C, et al. Whole genome sequencing detects inter-facility transmission of carbapenem-resistant Klebsiella pneumoniae. J Infect. 2019;78(3):187–199. doi:10.1016/j.jinf.2018.11.003
38. Choby JE, Howard-Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J Intern Med. 2020;287:283–300. doi:10.1111/joim.13007
39. Takeuchi D, Akeda Y, Yoshida H, et al. Genomic reorganization by IS26 in a blaNDM-5-bearing FII plasmid of Klebsiella pneumoniae isolated from a patient in Japan. J Med Microbiol. 2018;67:1221–1224. doi:10.1099/jmm.0.000817
40. Sun L, Xu J, He F. Draft genome sequence of an NDM-5, CTX-M-15 and OXA-1 co-producing Escherichia coli ST167 clinical strain isolated from a urine sample. J Glob Antimicrob Resist. 2018;14:284–286. doi:10.1016/j.jgar.2018.08.005
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



