Back to Journals » Infection and Drug Resistance » Volume 14
Epidemic Potential of Escherichia coli O16:H41-ST131: Compared with Pandemic O25b:H30-ST131 Lineage
Authors Zhang S, Zhang Q, Huang J, Cao Y, Zhao Z , Li B
Received 1 April 2021
Accepted for publication 1 June 2021
Published 8 July 2021 Volume 2021:14 Pages 2625—2632
DOI https://doi.org/10.2147/IDR.S313261
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Suresh Antony
Shengcen Zhang,1,* Qianwen Zhang,1,* Jiangqing Huang,1,* Yingping Cao,1 Zhichang Zhao,2 Bin Li1
1Department of Clinical Laboratory, Fujian Medical University Union Hospital, Fuzhou, Fujian, 350001, People’s Republic of China; 2Department of Pharmacy, Fujian Medical University Union Hospital, Fuzhou, Fujian, 350001, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhichang Zhao
Department of Pharmacy, Fujian Medical University Union Hospital, 29 Xinquan Road, Fuzhou, Fujian, 350001, People’s Republic of China
Email [email protected]
Bin Li
Department of Clinical Laboratory, Fujian Medical University Union Hospital, 29 Xinquan Road, Fuzhou, Fujian, 350001, People’s Republic of China
Email [email protected]
Background: O16:H41 is an important subclone among Escherichia coli (E. coli) sequence type (ST) 131, which has risen dramatically in recent years. However, reasons for the rapid increase of E. coli O16:H41-ST131 remain unclear. The aim of this study was to compare the pathogenicity and survivability features of E. coli O16:H41-ST131 with global epidemic O25b:H30-ST131 lineage.
Methods: Sixteen E. coli ST131 were divided into two groups: group O16:H41-ST131 (n=6) and group O25b:H30-ST131 (n=10). Adhesion and invasion activity of different isolates were measured using human T24 cells. Biofilm production was quantified by crystal violet staining. Fifty percent human serum was used to detect serum sensitivity. Resistance to hydrogen peroxide was detected by broth microdilution method, and anti-phagocytic function was determined by phagocytosis experiments.
Results: E. coli O16:H41-ST131 and O25b:H30-ST131 lineage showed similar biofilm formation, adhesion and invasion abilities. In terms of survivability, resistance to serum and hydrogen peroxide of E. coli O16:H41-ST131 was similar as that of E. coli O25b:H30-ST131. But anti-phagocytic function of E. coli O16:H41-ST131 was significantly weaker than that of E. coli O25b:H30-ST131.
Conclusion: The pathogenicity and survivability of E. coli O16:H41-ST131 were similar to those of E. coli O25b:H30-ST131, which may be important reasons for its increasing prevalence. Our study may contribute to a better understanding of the prevalence of E. coli O16:H41-ST131.
Keywords: O16:H41, O25b:H30, ST131, pathogenicity, survivability
Introduction
Escherichia coli (E. coli), which is commonly found in the gut flora of human beings and animals, can cause a wide range of infections such as septicemia, meningitis, and urinary tract infections.1 Sequence type (ST) 131, identified in 2008, is currently recognized as a predominant lineage among E. coli worldwide.1,2 Almost all of E. coli ST131 isolates are resistant to fluoroquinolones and commonly carry extended-spectrum β-lactamase genes like blaCTX-M-15.3 Therefore, the global increase in antibiotic resistance of E. coli is closely related to expansion of ST131 lineage.4 The epidemiological success of E. coli ST131 clonal group may be attributed to its enhanced pathogenicity and stronger survivability.5
O25b was generally considered as a predominant E. coli ST131 clonal serotype. However, the prevalence of E. coli O16-ST131 lineage significantly rose in recent years and made an important contribution to the expanding population of E. coli.4,6 A previous study showed that 33.7% of E. coli ST131 clinical isolates are O16 serotype in China.7,8 Besides, other research found that O16-ST131 lineage was a dominant lineage among fecal strains in China.8 In addition, the most prevalent lineage within E. coli ST131 is known as H30-ST131, which is often associated with many adverse consequences, such as multidrug-resistance (MDR) and persistent infections.9 Meanwhile, E. coli H41-ST131 is also a non-negligible lineage whose prevalence is even higher than that of E. coli H30-ST131 in certain regions.10,11
E. coli ST131 can be classified into different clonal groups that differ in their virulence and survivability.4 Therefore, characteristics of different lineages should be described separately when exploring the causes of their epidemics. Our previous study showed that O25b-ST131 lineage (66.4%) was a dominant clinical E. coli ST131 lineage, followed by O16-ST131 subclone (33.6%).7 Our results suggested that O16-ST131 lineage was poised to become a major serotype of E. coli ST131 in China. Therefore, the aim of this study was to assess the difference in pathogenicity and survivability features between E. coli O16:H41-ST131 and global epidemic O25b:H30-ST131 lineage for a more comprehensive understanding of the epidemic potential of E. coli O16:H41-ST131.
Materials and Methods
Bacterial Isolates
16 E. coli ST131, collected from Union Hospital of Fujian Medical University and reported in our previous study,7 were randomly selected in this study. 16 E. coli ST131 were divided into two groups, group O16:H41-ST131 (n=6) and group O25b:H30-ST131 (n=10).
Pathogenicity Analysis
Adhesion Assay
Adhesion abilities were detected using human bladder cancer cells (T24 cells, Anchorage-dependent cell, FH0171, FuHeng Cell Center, Shanghai, China), performed as previously described.12 Briefly, T24 cells were incubated with E. coli ST131 in 24-well plates at 37°C, 5% CO2 for 3 hours and infected with multiplicity of infection (MOI) of 10. Thereafter, T24 cells were washed with 1×phosphate-buffered saline (PBS) thrice. Then T24 cells were lysed with 0.1% Triton X-100 for 10 minutes. The lysates were diluted serially and plated on MH-agar plates for bacterial count. E. coli strain EC505 was used as positive control and E. coli DH5α served as negative control.
Invasion Assay
In invasion assay, the treatments of T24 cells were similar to those in adhesion assay but an additional step was carried out.12 After 3 hours of incubation, medium was replaced with fresh medium containing 100µg/mL gentamicin and incubated for 1.5 hours to kill extracellular strains. Then T24 cells were washed with 1×PBS thrice and lysed with 0.1% Triton X-100 for 10 minutes. The lysates were diluted serially and plated on MH-agar plates for bacterial count.
Biofilm Formation Experiments
Biofilm formation capacity was quantified by crystal violet assay based on a method described in previous studies with a few modifications.13,14 The strains were grown overnight in LB broth medium at 37°C under stationary aerobic conditions. Then, 0.5 McFarland turbidity standard saline-washed cultures were diluted 1:100 in LB broth medium and 200 μL cell suspensions were inoculated into 96 flat-bottomed well, polystyrene microtiter plates. The cultures were incubated for 48 h at 37°C without shaking. After incubation, each well was washed with phosphate-buffered saline (PBS) to remove planktonic bacteria and fresh 200 μL LB broth medium with or without ciprofloxacin (CIP) (2μg/mL) was added to each well. Following incubation for 24 h, each well was gently washed with 200 μL of phosphate-buffered saline (PBS) three times and stained with 200 μL of 1% crystal violet for 15 min at room temperature. Then, the plates were washed with distilled water to remove excess dye. To quantify biofilm production, 200 μL of anhydrous ethanol was used to solubilize crystal violet. The optical density was measured at 595 nm in ELISA reader. LB broth medium without bacterial cultures was used as negative control (ODc) and Enterococcus faecalis (E. faecalis) ATCC 29212 served as positive control.15 The degree of biofilm production was classified according to the following criteria: strong (OD>2×ODc), moderate (1.5×ODc<OD≤2×ODc), weak (ODc<OD≤1.5×ODc), absent (OD≤ODc). The concentration of CIP was defined according to bioavailability in human urine.16
Viability Analysis
Serum Sensitivity Test
To analyze serum sensitivity, strains were incubated with 50% human serum as described previously.12 Resistance to serum was determined by the number of colonies (CFU/mL) recovered from each well.12 E. coli strain EC505 was used as positive control and E. coli DH5α served as negative control.
Hydrogen Peroxide Susceptibility Test
Hydrogen peroxide susceptibility was detected by broth microdilution method, according to the Clinical and Laboratory Standards Institute (CLSI, 2020) standards. Results were presented in the form of minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC). E. coli ATCC 25922 was used as quality control.
Phagocytosis Experiment
Anti-phagocytic functions were determined using RAW264.7 cells (Anchorage-dependent cell, FH0328, FuHeng Cell Center, Shanghai, China), performed as previously described.17 Briefly, 2×105 /mL RAW264.7 cells and 2×106 CFU/mL bacteria were incubated at 37°C for 1.5 hours. Trypan blue staining cell viability assay kit was used to determine viability of RAW264.7 cells. When phagocytosed, E. coli can be easily recognized under a microscope by Gram staining, which allows explicit and quantitative measurement of cell phagocytosis. Therefore, microscopy was used to visualize phagocytosed E. coli. Anti-phagocytic functions were observed in terms of anti-phagocytosis rate (PR): PR = (total numbers of cells harbored the phagocytosed E. coli in 200 cells)/200×100%. E. coli ATCC 25922 was used as positive control and E. coli DH5α served as negative control. All experiments were conducted three times in three replicates.
Statistical Analysis
The Mann–Whitney U test was performed to assess differences in adhesion capabilities, invasion abilities, serum sensitivities, hydrogen peroxide susceptibilities, and biofilm formation capabilities between two groups. The two-sample t-test was conducted to evaluate the variation in anti-phagocytic function between two groups (E. coli O25b:H30-S131 and E. coli O16:H41-ST131) and the relationship between biofilm formation and antibiotics. All analyses were performed in SPSS-25 with a significance level of α = 0.05.
Results
Pathogenic Potentials
Adhesion and Invasion Capabilities
Our study showed that adhesion and invasion abilities of two groups were significantly higher than those of E. coli DH5α (Figure 1, p <0.05). But the results showed no significant difference in adhesion and invasion abilities between two groups (E. coli O25b:H30-S131 and E. coli O16:H41-ST131) (Figure 1, p <0.05).
Biofilm Formation Capability
As quantified by crystal violet staining, all isolates were able to form biofilms in LB broth medium with or without CIP. Among them, 83.33% of E. coli O16:H41-ST131 and 70% of E. coli O25b:H30-ST131 showed weak biofilm forming ability. Although 83.33% of E. coli O16:H41-ST131 still presented weak biofilm forming ability, CIP significantly reduced biofilm biomass of 50% of E. coli O16:H41-ST131 (Table 1). In group O25b:H30-ST131, strong biofilm forming strains EC549 showed a significant reduction in biofilm forming ability (t=19.067, p<0.05) and it showed weak biofilm forming ability in LB broth with CIP. There was no significant difference between two groups (E. coli O25b:H30-S131 and O16:H41-ST131 lineage) in LB broth medium with or without CIP observed (p>0.05). The results of biofilm formation assay were shown in Table 1.
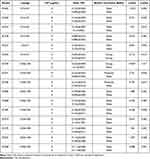 |
Table 1 Biofilm Formation Capability Among Different E. coli-ST131 Isolates |
Survival Capacity
Serum Sensitivity
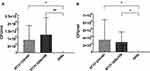
In our study, serum sensitivity was detected using 50% human serum. E. coli O16:H41-ST131 and E. coli O25b:H30-ST131 showed significantly higher resistance to serum than E. coli DH5α (Figure 2A, p<0.05). But two groups showed similar resistance to human serum (p>0.05).
Susceptibility to Hydrogen Peroxide
In this study, the MIC value of hydrogen peroxide for all E. coli ST131 was 16 μg/mL or higher. The MIC and MBC values of hydrogen peroxide for E. coli ATCC 25922 were all 8 μg/mL (Table 2). It was observed that there were significant differences between E. coli O16:H41-ST131 and E. coli ATCC 25922 both in MIC and MBC. But no significant difference in resistance to hydrogen peroxide was observed between two groups of isolates (E. coli O16:H41-ST131 and E. coli O25b:H30-ST131 lineage) at MIC level (Figure 2B) and MBC level (Figure 2C).
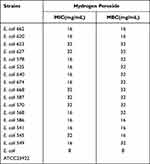 |
Table 2 Antibacterial Activity of Hydrogen Peroxide Against E. coli O16:H41-ST131 and E. coli O25b:H30-ST131 |
Anti-Phagocytic Function
The results of anti-phagocytic function assay were shown in Figure 2D. The phagocytosis of E. coli O16:H41-ST131 by RAW264.7 cells was significantly lower than that of E. coli DH5α (t=7.418, p<0.001). Meanwhile, anti-phagocytic function of E. coli O16:H41-ST131 was significantly weaker than that of E. coli O25b:H30-ST131 (t=−2.527, p<0.05).
Discussion
Within the E. coli population, ST131 is a dominant MDR lineage worldwide.4 Recent studies showed that ST131 lineage carried more virulence genes, compared with other E. coli lineages.18 It was conventionally assumed that O25b was a predominant ST131 serotype, but the prevalence of E. coli O16-ST131 significantly rose in recent years.7,8 Different E. coli ST131 clone groups possess various virulence and survivability features.15 Therefore, in order to further explore the reasons for increased prevalence of E. coli O16:H41-ST131, we analyzed the difference in pathogenicity and survivability between E. coli O16:H41-ST131 and global epidemic O25b:H30-ST131 lineage.
In this study, adhesion, invasion and biofilm formation capabilities were used to assess pathogenicity. Previous studies suggested that adhesion and invasion capabilities seemed to be the key of bacterial pathogenesis and important factors of some adverse events such as antibiotic resistance and bacterial persistence.19,20 Our study showed that both groups of isolates (E. coli O16:H41-ST131 and E. coli O25b:H30-ST131) exhibited strong adhesion and invasion ability to human bladder cancer cells, consistent with previous studies21 (Figure 1). At the same time, our study found that E. coli O16:H41-ST131 and pandemic E. coli O25b:H30-ST131 exhibited similar adhesion and invasion abilities (Figure 1, p>0.05). This result suggested that E. coli O16:H41-ST131, like O25b:H30, might be able to colonize and establish infections when they enter human tissue. Occurrence of infections and enhanced antibiotic resistance are also closely related to the biofilm formation capacity of bacteria.22 It is hard for antibiotics to penetrate the biofilm, so the bacteria in biofilm are easy to become tolerant and resistant to antibiotics.23 Meanwhile, it was found that weak biofilm forming strains induced weaker immune responses than strong biofilm forming strains, possibly leading to immune evasion.24 In our study, we found that all of E. coli ST131 were able to form biofilms and the majority of E. coli O25b:H30-ST131 (70%) and E. coli O16:H41-ST131 (83.3%) presented (p>0.05) weak biofilm formation ability, consistent with previous studies.25 All strains used in our study were resistant to fluoroquinolones, so we also assessed their biofilm formation capacity post addition of CIP. Although CIP produced a significant reduction in biofilm biomass of 50% of E. coli O16:H41-ST131 and 30% of E. coli O25b:H30-ST131, all strains were still able to form biofilms.
Our results suggested that refractory infections caused by E. coli O16:H41-ST131 might be related to its biofilm formation capacity.
Strong survivability is also a contributory factor to prevalence of pathogenic bacteria. In our study, serum resistance, hydrogen peroxide resistance, and anti-phagocytic function were used to assess survivability. Serum resistance and anti-phagocytic function were considered as important determinants for survival of bacteria in vivo.12,26 Our study found that E. coli O16:H41-ST131 was capable of growing in human serum, indicating strong serum resistance capacity of this lineage. Serum resistance is one of the important mechanisms of bacteria enabling them to survive in the bloodstream of the host.12 We supposed that the strong serum resistance might contribute to the current increase in bloodstream infections caused by E. coli O16:H41-ST131. Besides, our study illustrated that anti-phagocytic function of E. coli O16:H41-ST131 was lower than that of E. coli O25b:H30-ST131, but still significantly higher than that of E. coli DH5α (Figure 2, p<0.05). Anti-phagocytic function of bacteria plays an important role in the occurrence of infections. Strong anti-phagocytic function can protect E. coli strains from phagocytosis and ensure their survival in vivo, which conduces to causing infections. These results suggested that not only pandemic E. coli O25b:H30-ST131 had resistance to bactericidal effect of human immune system, but E. coli O16:H41-ST131 could resist human immune function and cause infections. Resistance to hydrogen peroxide, often used as disinfectant in hospitals, seems to be closely related to hospital-related bacterial infections.27 Two groups of strains (E. coli O16:H41-ST131 and E. coli O25b:H30-ST131) showed similar hydrogen peroxide resistance, which was significantly higher than that of E. coli ATCC 25922 (Figure 2, p<0.05). As discussed previously, our results proved that E. coli O16:H41-ST131 and epidemic E. coli O25b:H30-ST131 showed similar pathogenicity and survivability features, which might contribute to the epidemiological success of E. coli O16:H41-ST131.
However, there are some limitations to our study which should be considered in further research to better understand the reasons for increasing prevalence of E. coli O16:H41-ST131. First, the sample size of E. coli strains O16:H41-ST131 (n=6) and group O25b:H30-ST131 (n=10) in our study was small. All strains were collected from the same hospital so they might have a genetic relationship. Therefore, multi-center research with a large sample size should be conducted in our future studies for a more comprehensive understanding of survivability and pathogenicity of ST131. Second, our study only assessed the number of phagocytosed bacteria, but did not detect the survival of bacteria in RAW 264.7 cells. The survival of bacteria in RAW 264.7 cells might contribute to a better understanding of the pathogenesis of E. coli O16:H41-ST131 in vivo, so further research is still needed.
Conclusion
In this study, the pathogenicity and survivability features of E. coli O16:H41-ST131 were analyzed. The pathogenicity and survivability of E. coli O16:H41-ST131 were similar to those of pandemic O25b:H30-ST131 lineage. But in terms of anti-phagocytic function, E. coli O16:H41-ST131 was slightly inferior to O25b:H30-ST131 lineage. Our research could contribute to a comprehensive understanding of the increasing prevalence of E. coli O16:H41-ST131 and suggests that it should be continuously monitored to cope with the increase in infections caused by E. coli ST131.
Abbreviations
ST, sequence type; CIP, ciprofloxacin; MIC, minimum inhibitory concentration; MBC, minimum bactericidal concentration; PR, anti-phagocytosis rate.
Ethics Approval and Informed Consent
All procedures of this study involving humans (individuals, medical records, human samples, clinical isolates and human cell lines) were reviewed and approved by the Medical Ethics Committee of Fujian Medical University Union Hospital (2020KY0121). All the patients participating in this study signed informed consent, while the guardians of children aged less than 18 years signed on behalf of them. We confirm that this study was conducted in accordance with the Declaration of Helsinki.
Consent for Publication
All authors confirm that the details of any images, videos, recordings, etc can be published.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Joint Funds for the innovation of science and Technology, Fujian province [Grant number: 2017Y9049] and the Educational and Scientific Research Project for Young and Middle-Aged Teachers of Fujian Province (Grant number: JAT190191).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Manges AR, Geum HM, Guo A, Edens TJ, Fibke CD, Pitout JDD. Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin Microbiol Rev. 2019;32(3):e00135–18. doi:10.1128/CMR.00135-18
2. Fibke C, Croxen M, Geum H, et al. Escherichia coli genomic epidemiology of major extraintestinal pathogenic lineages causing urinary tract infections in young women across Canada. Open Forum Infect Dis. 2019;6(11):ofz431. doi:10.1093/ofid/ofz431
3. Johnson J, Tchesnokova V, Johnston B, et al. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis. 2013;207(6):919–928. doi:10.1093/infdis/jis933
4. Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27(3):543–574. doi:10.1128/CMR.00125-13
5. Ranjan A, Shaik S, Hussain A, et al. Genomic and functional portrait of a highly virulent, CTX-M-15-producing H30-Rx subclone of Escherichia coli sequence type 131. Antimicrob Agents Chemother. 2015;59(10):6087–6095. doi:10.1128/AAC.01447-15
6. Mathers A, Peirano G, Pitout J. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28(3):565–591. doi:10.1128/CMR.00116-14
7. Li B, Lu Y, Lan F, He Q, Li C, Cao Y. Prevalence and characteristics of ST131 clone among unselected clinical Escherichia coli in a Chinese university hospital. Antimicrob Resist Infect Control. 2017;6(1):118. doi:10.1186/s13756-017-0274-0
8. Zhong YM, Liu WE, Liang XH, Li YM, Jian ZJ, Hawkey PM. Emergence and spread of O16-ST131 and O25b-ST131 clones among faecal CTX-M-producing Escherichia coli in healthy individuals in Hunan Province, China. J Antimicrob Chemother. 2015;70(8):2223–2227. doi:10.1093/jac/dkv114
9. Gibreel T, Dodgson A, Cheesbrough J, Fox A, Bolton F, Upton MJT. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother. 2012;67(2):346–356. doi:10.1093/jac/dkr451
10. Rogers BA, Ingram PR, Runnegar N, et al. Sequence type 131 fimH30 and fimH41 subclones amongst Escherichia coli isolates in Australia and New Zealand. Int J Antimicrob Agents. 2015;45(4):351–358. doi:10.1016/j.ijantimicag.2014.11.015
11. Ho PL, Chu YP, Lo WU, et al. High prevalence of Escherichia coli sequence type 131 among antimicrobial-resistant E. coli isolates from geriatric patients. J Med Microbiol. 2015;64(Pt 3):243–247. doi:10.1099/jmm.0.000012
12. Shaik S, Ranjan A, Tiwari SK, et al. Comparative genomic analysis of globally dominant ST131 clone with other epidemiologically successful extraintestinal pathogenic Escherichia coli (ExPEC) lineages. MBio. 2017;8(5):e01596–01517. doi:10.1128/mBio.01596-17
13. Wang Y, Yi L, Wang Y, et al. Isolation, phylogenetic group, drug resistance, biofilm formation, and adherence genes of Escherichia coli from poultry in central China. Poult Sci. 2016;95(12):2895–2901. doi:10.3382/ps/pew252
14. Gonzalez MJ, Robino L, Iribarnegaray V, Zunino P, Scavone P. Effect of different antibiotics on biofilm produced by uropathogenic Escherichia coli isolated from children with urinary tract infection. Pathog Dis. 2017;75(4):ftx053. doi:10.1093/femspd/ftx053
15. Cieśla J, Stępień-Pyśniak D, Nawrocka A, et al. Surface properties of Enterococcus faecalis cells isolated from chicken hearts determine their low ability to form biofilms. Biofouling. 2018;34(2):149–161. doi:10.1080/08927014.2017.1416105
16. Lorian V, editor. Antibiotics in Laboratory Medicine. Lippincott Williams & Wilkins; 2005.
17. Zhu H, Yan L, Gu J, Hao W, Cao J. Kv1.3 channel blockade enhances the phagocytic function of RAW264.7 macrophages. Sci China Life Sci. 2015;58(9):867–875. doi:10.1007/s11427-015-4915-3
18. Alqasim A, Abu Jaffal A, Alyousef AJ. Escherichia coli prevalence and molecular characteristics of sequence type 131 clone among clinical uropathogenic isolates in Riyadh, Saudi Arabia. Saudi J Biol Sci. 2020;27(1):296–302. doi:10.1016/j.sjbs.2019.09.020
19. Chakroun I, Cordero H, Mahdhi A, et al. Adhesion, invasion, cytotoxic effect and cytokine production in response to atypical Salmonella Typhimurium infection. Microb Pathog. 2017;106:40–49. doi:10.1016/j.micpath.2016.11.004
20. Hussain A, Shaik S, Ranjan A, et al. Genomic and functional characterization of poultry escherichia coli from india revealed diverse extended-spectrum β-lactamase-producing lineages with shared virulence profiles. Front Microbiol. 2019;10:2766. doi:10.3389/fmicb.2019.02766
21. Ramos NL, Sekikubo M, Dzung DT, et al. Uropathogenic Escherichia coli isolates from pregnant women in different countries. J Clin Microbiol. 2012;50(11):3569–3574. doi:10.1128/JCM.01647-12
22. Beebout C, Eberly A, Werby S, et al. Respiratory heterogeneity shapes biofilm formation and host colonization in uropathogenic Escherichia coli. MBio. 2019;10(2):e02400–18. doi:10.1128/mBio.02400-18
23. Verderosa A, Totsika M, Fairfull-Smith K. Bacterial biofilm eradication agents: a current review. Front Chem. 2019;7:824. doi:10.3389/fchem.2019.00824
24. Gogoi-Tiwari J, Williams V, Waryah CB, et al. Comparative studies of the immunogenicity and protective potential of biofilm vs planktonic Staphylococcus aureus vaccine against bovine mastitis using non-invasive mouse mastitis as a model system. Biofouling. 2015;31(7):543–554. doi:10.1080/08927014.2015.1074681
25. Sarkar S, Vagenas D, Schembri M, Totsika MJP. Biofilm formation by multidrug resistant Escherichia coli ST131 is dependent on type 1 fimbriae and assay conditions. Pathog Dis. 2016;74(3):ftw013. doi:10.1093/femspd/ftw013
26. Xu B, Zhang P, Zhou H, Sun Y, Tang J, Fan H. Identification of novel genes associated with anti-phagocytic functions in Streptococcus equi subsp. zooepidemicus. Vet Microbiol. 2019;233:28–38. doi:10.1016/j.vetmic.2019.04.023
27. Alotaibi S, Ayibiekea A, Pedersen A, et al. Susceptibility of vancomycin-resistant and -sensitive Enterococcus faecium obtained from Danish hospitals to benzalkonium chloride, chlorhexidine and hydrogen peroxide biocides. J Med Microbiol. 2017;66(12):1744–1751. doi:10.1099/jmm.0.000642
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.