Back to Journals » OncoTargets and Therapy » Volume 12
Emerging roles of circular RNAs in colorectal cancer
Authors Hao S, Cong L, Qu R, Liu R, Zhang G , Li Y
Received 10 March 2019
Accepted for publication 14 May 2019
Published 19 June 2019 Volume 2019:12 Pages 4765—4777
DOI https://doi.org/10.2147/OTT.S208235
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Federico Perche
Shuhong Hao,1 Liang Cong,2 Rongfeng Qu,1 Rui Liu,1 Guizhen Zhang,3 Yarong Li1
1Department of Hematology and Oncology, The Second Hospital of Jilin University, Changchun, Jilin, People’s Republic of China; 2Department of Digestive Endoscopy, The Second Hospital of Jilin University, Changchun, Jilin, People’s Republic of China; 3Department of Orthopedics, The Second Hospital of Jilin University, Changchun, Jilin, People’s Republic of China
Abstract: Circular RNAs (circRNAs) are a newly discovered class of endogenous non-coding RNAs. Owing to the development of high-throughput sequencing, researchers have identified thousands of circRNAs. Emerging evidence suggests that circRNAs are involved in various tumor cell processes, including proliferation, apoptosis, invasion and migration. Because of their high stability and abundance, tissue-specific expression, and easy detection, circRNAs are considered ideal biomarkers for cancer diagnosis and prognosis. An increasing number of studies have recently demonstrated that circRNAs are closely associated with colorectal cancer (CRC). CRC is the third most common cancer and the second leading cause of cancer-related death globally. Thus, understanding the molecular mechanisms involved in the development and progression of CRC is vital. In this review, we summarize the current literature regarding human circRNAs related to CRC and present an overview of the potential clinical implications of circRNAs with respect to CRC.
Keywords: circRNA, biomarker, diagnosis, prognosis
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related death in men and women worldwide.1 Owing to population aging and improved standards of living, the global incidence of CRC has increased annually.2 In the United States, an estimated 140,250 cases of CRC were diagnosed in 2018, and 50,630 patients died of the disease.3 Studies have shown that several genetic factors are involved in CRC development, including changes in chromosomal copy number, aberrant gene methylation, and dysregulated gene expression.4 However, the potential molecular mechanisms contributing to CRC development and progression are still far from being fully understood.5 Non-metastatic colon cancer is generally treated by surgical colectomy combined with chemotherapy.6 With the development of new technologies for CRC treatment, the prognosis of patients who are diagnosed at early stages has improved.7 However, many CRC patients are diagnosed at advanced stages owing to the lack of incipient symptoms and limitations of timely screening methods, which greatly contribute to the poor prognosis of CRC.8 According to the statistics, the 5-year survival rates for patients with early-stage, localized disease (stages I and II) approach 90%, whereas the survival rate for those diagnosed with late-stage CRC, which is associated with distant metastasis, is 13.1%.9 Therefore, it is necessary to gain a deeper understanding of the molecular mechanisms involved in CRC progression as well as identify new diagnostic and prognostic biomarkers.
Circular RNAs (circRNAs) are a newly discovered class of endogenous non-coding RNAs.10 They were initially regarded as the results of mis-splicing or as byproducts of pre-mRNA processing at low abundance.11 Owing to the development of high-throughput sequencing, investigators have identified thousands of circRNAs in viruses, fungi, plants, and animals.12 CircRNAs are conserved, stable, and highly abundant and display species-, tissue-, and developmental stage-specific expression.13,14 In contrast to linear RNAs, circRNAs have a covalently closed continuous loop structure without 5′ caps or 3′ polyadenylated tails, and they are not sensitive to digestion by RNases; thus, circRNAs are more conserved and stable than linear RNAs.15,16 Based on their mechanism of formation, circRNAs can be divided into three categories, namely exonic circRNAs (EcircRNAs), intronic circRNAs (CiRNAs), and exon-intron circRNAs (EIciRNAs).17 Some functions of these circRNAs have been revealed: EcircRNAs, which encompass the majority of circRNAs, are predominantly cytoplasmic and may serve as microRNA (miRNA) sponges to enhance the expression levels of miRNA target genes.18 Intron-containing circRNAs, including CiRNAs and EIciRNAs, are abundant in the nucleus and may regulate the expression of their host genes.19 For example, ci-ankrd52 promotes the transcription of its host gene (ANKRD52) by modulating the elongation activity of Pol II.20 In addition, a few circRNAs were reported to have the potential to be translated into proteins. CircFBXW7, produced from the F-box and WD repeat domain containing 7 (FBXW7) gene, inhibited the proliferation and cell cycle acceleration of gliomas by translation into a functional protein named FVXW7-185aa.21 Moreover, circRNAs can function as competing endogenous RNAs (ceRNAs) by regulating cancer-related signaling pathways, such as the epidermal growth factor receptor (EGFR)/signal transducer and activator of transcription 3 (STAT3),22 phosphatidylinositol 3 kinase (PI3K)/protein kinase B (AKT),23 and Wnt/β-catenin pathways.24 Taken together, these findings indicate that circRNAs have great potential for use in regulation of biological processes and disease progression.
Evidence has indicated that circRNAs are involved in various physiological and pathological processes of tumor cells, including proliferation, apoptosis, invasion, and migration.25 For instance, circAGFG1 may act as a miRNA sponge of miR-195-5p to relieve the repressive effect of miR-195-5p on its target gene, cyclin E1 (CCNE1), thus promoting the proliferation, mobility, and invasion of triple-negative breast cancer cells in vivo.26 CircPVT1 serves as a ceRNA of miR-497 and indirectly regulates Bcl-2 expression in non-small cell lung cancer cells, thereby promoting the progression of non-small cell lung cancer.27 Importantly, circRNAs are stably expressed in saliva, blood, and exosomes, which makes them promising biomarkers for the diagnosis, prognosis, and therapeutic assessment of cancer patients.28 Using circRNA microarray analysis, Li et al identified 343 circRNAs that were differentially expressed in the plasma of gastric cancer patients versus healthy controls. Reverse transcription-droplet digital PCR (RT-ddPCR) was used to confirm that two of these circRNAs, hsa_circ_0001017 and hsa_circ_0061276, were downregulated in the plasma of gastric cancer patients compared with that in healthy controls, and these circRNA levels were significantly associated with tumor size, TNM stage, distal metastasis, and overall survival (OS).29 Further, Li et al revealed that circRNAs were more enriched in exosomes than in the producer cells, including liver, colon, lung, stomach, breast, and cervical cancer cell lines. In addition, they found that serum circRNAs contained in exosomes may distinguish patients with colon cancer from healthy controls, which illustrates their potential as circulating biomarkers for cancer diagnosis.30 However, the potential mechanisms and biological functions of most circRNAs in CRC have remained unclear and have therefore been a focus of investigation. In this review, we summarize the recent studies focused on human circRNAs involved in CRC and present an overview of the potential clinical implications of circRNAs in CRC.
Expression of circRNAs in CRC
With the development of high-throughput sequencing, extensive research has shown that circRNAs are differentially expressed in many cancers, including CRC. Utilizing circRNA chips, Chen et al identified 10,245 differentially expressed circRNAs in CRC tissues versus adjacent non-tumor tissues, including 6,264 upregulated and 3,981 downregulated circRNAs.31 By differential gene expression analysis of normal colon mucosa and CRC tissues, Bachmayr-Heyda et al revealed 39 significantly differentially expressed circRNAs; 11 were upregulated, and 28 were downregulated in CRC tissues versus adjacent non-tumor tissues.32 Similarly, Zhang et al discovered 23 differentially expressed circRNAs in CRC tissues.33 In addition, Yan et al screened the circRNA expression profiles in CRC and adjacent normal tissues using circRNA microarray analyses. Their results showed that 265 circRNAs were differentially expressed, among which 99 were upregulated and 166 were downregulated in CRC tissues. Based on their relationship with protein-coding genes, the dysregulated circRNAs were classified into five categories: 75.85% were exonic, 11.32% were intronic, 7.92% were overlapping, 1.51% were intergenic, and 3.4% were antisense.34 By comparing three isogenic CRC cell lines with different KRAS mutation states, including DLD-1, DKO-1, and DKs-8, Dou et al found that circRNAs were significantly downregulated at a global level in the KRAS-mutated cells compared with that in KRAS-wild-type cells, indicating that circRNAs are involved in KRAS mutations and may be potential biomarkers for KRAS-mutated CRC.35 Jiang et al examined the circRNAs in three cell lines, including primary CRC cells (SW480), metastatic CRC cells (SW620), and normal human colon mucosal epithelial cells (NCM460). They identified 2,919 differentially expressed circRNAs in CRC cells when compared with NCM460 cells. In addition, they revealed 623 differentially expressed circRNAs between SW480 and SW620 cells, thereby suggesting that these circRNAs are involved in CRC development and metastasis.36
circRNAs regulate the proliferation and progression of CRC
Accumulating evidence has indicated that circRNAs function as either oncogenes or tumor suppressors by regulating the proliferation, invasion, migration, and apoptosis of CRC cells. Various mechanisms, such as miRNA sponging, peptide translation, and cancer-related signaling pathway regulation, are involved in these functions. Of these, miRNA sponging is the main mechanism of circRNAs observed in CRC cells. The functions and mechanisms of dysregulated circRNAs in CRC are shown in Table 1.
 | Table 1 CircRNAs regulate the proliferation and progression of colorectal cancer |
CircRNAs act as oncogenes in CRC
CircACAP2
CircACAP2, also known as hsa_circ_0007331, was reported to be significantly upregulated in CRC tissues and colon cancer SW480 cells compared with that in normal controls. Knockdown of circACAP2 inhibited the proliferation, migration, and invasion of SW480 cells. Furthermore, circACAP2 silencing promoted the expression of miR-21-5p, which in turn inhibited the expression of T lymphoma invasion and metastasis protein 1 (Tiam1) at the mRNA and protein level, demonstrating that circACAP2 regulates Tiam1 in CRC by sponging miR-21-5p.37
Hsa_circ_0071589
Hsa_circ_0071589, which is located on chr4:187517693–187518946, has a length of 743 bp, and is produced from the FAT atypical cadherin 1 (FAT1) gene,38 was found to be significantly overexpressed in CRC tissues compared with that in normal colon tissues. In CRC cells, blockage of hsa_circ_0071589 inhibited tumor growth, invasion, and migration. Through bioinformatics analysis, luciferase reporter assay, and RNA immunoprecipitation (RIP) assay, hsa_circ_0071589 was shown to function as a sponge of miR-600, and enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) was identified as a target gene of miR-600.39 These results suggest that hsa_circ_0071589 promotes carcinogenesis via the miR-600/EZH2 axis in CRC.
Hsa_circ_0007534
Hsa_circ_0007534 is an exon-derived circRNA that originates from the protein-coding gene DEAD-box helicase 42 (DDX42).38 It was found to be upregulated in a variety of cancers, including glioma,40 osteosarcoma,41 breast cancer,42 non-small cell lung cancer,43 and CRC.44 Li et al found that hsa_circ_0007534 promoted glioma cell proliferation and migration via upregulating Zic family member 5 (ZIC5) expression by sponging miR-761.40 In addition, Li et al found that hsa_circ_0007534 promoted growth and restrained apoptosis in osteosarcoma cells by affecting the AKT/GSK-3β signaling pathway.41 Song et al found that knockdown of hsa_circ_0007534 significantly suppressed progression in breast cancer cells by reducing miR-593, which promoted mucin 19 (MUC19) production.42 Zhang et al detected hsa_circ_0007534 levels in cancer tissues and adjacent non-tumorous tissues from 33 CRC patients and demonstrated that expression of hsa_circ_0007534 was significantly upregulated in the CRC tissues. Moreover, hsa_circ_0007534 expression was correlated with tumor stage and lymph node metastasis. Furthermore, silencing of hsa_circ_0007534 by siRNA significantly inhibited proliferation and induced apoptosis of CRC cells.44 However, the functional mechanisms of hsa_circ_0007534 in CRC require further investigation.
Hsa_circ_001569
Hsa_circ_001569 is produced from the ATP binding cassette subfamily C member 1 (ABCC1) gene.38 Xie et al found that hsa_circ_001569 was upregulated in CRC tissues, and its expression level was positively correlated with TNM stage and poor differentiation.45 MiR-145, as a tumor suppressor, was found to inhibit cell proliferation and invasion in several cancers, such as non-small cell lung cancer,46 intrahepatic cholangiocarcinoma,47 and colon cancer.48 Hsa-circ_001569 was demonstrated to promote proliferation and invasion by directly inhibiting miR-145 transcription and subsequently affecting the functions of the miR-145 targets, E2F transcription factor 5 (E2F5), BCL2 associated athanogene 4 (BAG4), and formin like 2 (FMNL2), in CRC cells.45
Hsa_circ_000984
Hsa_circ_000984 is 725 bp and generated from the cyclin-dependent kinase 6 (CDK6) gene.38 CDK6 is a key factor in cell cycle regulation and is essential for passage into the G1 phase.49 A study by Xu et al revealed that hsa_circ_000984 is markedly overexpressed in CRC patients and CRC cell lines. Further analysis revealed that hsa_circ_000984 knockdown inhibited CRC cell proliferation, migration, and invasion in vitro and tumor formation in vivo. Mechanistically, hsa_circ_000984 was found to act as a ceRNA by binding miR-106b and effectively upregulating the expression of CDK6, which is a target of miR-106b.50
Hsa_circ_0020397
Hsa_circ_0020397 is an exon-derived circRNA that originates from the protein-coding gene dedicator of cytokinesis 1 (DOCK1).38 Compared with that in normal colonic epithelial cells, hsa_circ_0020397 was significantly overexpressed in CRC cells.51 MiR-138, a well-known tumor suppressor that is downregulated in a variety of cancers,51–53 was confirmed to be a target of hsa_circ_0020397 by dual-luciferase reporter assay.51 Functional studies revealed that hsa_circ_0020397 regulated the viability and apoptosis of CRC cells by promoting the expression of the miR-138 targets telomerase reverse transcriptase (TERT) and programmed death-ligand 1 (PD-L1).51 Tumor immune escape is an important strategy for tumor survival, and immunotherapy has been a cancer research hotspot in recent years. The interaction between PD-L1 and programmed death-1 (PD-1) is an important component of tumor immune escape; this interaction can inhibit the activation of T lymphocytes and enhance the immune tolerance of tumor cells, thereby achieving tumor immune escape.54 In addition to regulating the expression of PD-L1, accumulating evidence suggests that circRNAs play various roles in the modulation of antitumor immune responses. By decreasing miR-17-5p, Circ-Amotl1 increased the expression and facilitated the nuclear translocation of signal transducer and activator of transcription (Stat3),55 which is crucial for suppression of antitumor immunity in the tumor microenvironment.56 Moreover, the complex of circFoxo3 and MDM2 induces p53 degradation.57 In tumor stromal cells, p53 plays a key role in tumor development by inducing an increase in inducible nitric oxide synthase expression and modulating immune response.58 Based on these data, circRNAs can be considered novel targets in cancer immunotherapy. However, the underlying mechanisms of circRNAs in cancer remain largely unknown, warranting further research.
CircBANP
CircBANP, generated from exons 5–11 of BTG3 associated nuclear protein (BANP), was originally identified as a significantly overexpressed circRNA in CRC tissues by microarray, which was later verified by qRT-PCR. Knockdown of circBANP with siRNA repressed the proliferation of CRC cells and reduced the phosphorylation of Akt, suggesting that the Akt pathway is involved in circBANP-induced cell proliferation.59 Recently, upregulated expression of circBANP was also observed in lung cancer.60 In contrast to its function of Akt pathway regulation in CRC, circBANP was found to promote lung cancer growth, migration, and invasion by regulating the miR-503/La ribonucleoprotein domain family member 1 (LARP1) axis.60
Hsa_circ_0055625
Using microarray analysis, Zhang et al identified 198 upregulated and 136 downregulated circRNAs in colon cancer tissues. Among the top 10 upregulated circRNAs, hsa_circ_0055625 was confirmed to be significantly upregulated in colon cancer tissues by qRT-PCR. Moreover, the expression of hsa_circ_0055625 was found to be associated with pathological TNM stage and metastasis, indicating that hsa_circ_0055625 is a potential oncogenic biomarker of colon cancer. Further in vitro studies revealed that hsa_circ_0055625 regulated CRC cell proliferation and metastasis via the miR-106/integrin subunit beta 8 (ITGB8) axis.61
Hsa_circ_0000069
The expression of hsa_circ_0000069 was upregulated in 30 CRC tissues compared with that in paired noncancerous tissues. Expression of hsa_circ_0000069 was positively correlated with age and TNM stage. Functionally, hsa_circ_0000069 knockdown using siRNAs inhibited cell proliferation, migration, and invasion and induced G0/G1 phase arrest.62
circRNAs act as tumor suppressors in CRC
circITCH
As a well-known circRNA, circITCH has been reported to act as a sponge of oncogenic miR-7, miR-17, and miR-214 to enhance itchy E3 ubiquitin protein ligase (ITCH) expression in lung cancer63 and esophageal squamous cell carcinoma.64 Huang et al detected circITCH levels in cancer tissues and adjacent non-tumorous tissues in 45 CRC patients and demonstrated that the expression of circITCH was significantly downregulated in the CRC tissues. Their follow-up experiments showed that, by sponging miR-7 and miR-20a, circITCH increased the level of ITCH, which is involved in inhibition of the Wnt/β-catenin pathway.24
Hsa_circ_0000523
Hsa_circ_0000523 was reported to be downregulated in 12 CRC cell lines, including DLD-1, HCT-15, HT-29, and SW480. Interference of hsa_circ_0000523 induced the proliferation and suppressed apoptosis of CRC cells by upregulating miR-31, which exerted its tumor-promoting effects by downregulating dickkopf WNT signaling pathway inhibitor 1 (Dkk1), an antagonist of Wnt signaling.65 These results suggest that low levels of hsa_circ_0000523 lead to activation of the Wnt/β-catenin signaling pathway, subsequently inducing CRC progression.
Hsa_circ_103809
Hsa_circ_103809, also known as circZFR, has been shown to be overexpressed in lung cancer tissues, and knockdown thereof with siRNA repressed the proliferation of lung cancer cells and delayed tumor growth in vivo.66 However, hsa_circ_103809 plays an opposite role in CRC, in which its expression is downregulated in both tissues and cells. Functionally, downregulation of hsa_circ_103809 was shown to promote the proliferation and migration of CRC cells by regulating the miR-532-3P/forkhead box O4 (FOXO4) axis.67
circDDX17
Via RNA sequencing, Li et al identified 448 differentially expressed circRNAs in CRC, including 394 significantly upregulated and 54 significantly downregulated circRNAs. Using qRT-PCR, circDDX17 was confirmed to be significantly downregulated in 60 CRC tissues compared with that in paired normal tissues. In vitro experiments revealed that silencing of circDDX17 promoted CRC cell proliferation, invasion, and migration and inhibited apoptosis.68
circRNAs may serve as potential diagnostic and prognostic biomarkers for CRC
Because circRNAs are easier to extract and detect compared with proteins,69 they are ideal biomarkers for cancer diagnosis and prognosis. Based on their expression in CRC, we classified circRNAs as biomarkers that are upregulated or downregulated in CRC (Table 2).
 | Table 2 CircRNAs represent potential diagnostic and prognostic biomarkers for colorectal cancer |
Upregulated circRNAs in CRC
CiRS-7
CiRS-7, also known as CDR1as or hsa_circ_0001946, is the most well-known circRNA. CiRS-7 acts as an oncogenic circRNA to promote the progression of some cancers, such as gastric cancer,23 non-small cell lung cancer,70,71 esophageal squamous cell carcinoma,72,73 laryngeal cancer,74 and hepatocellular carcinoma,75 by functioning as a miR-7 sponge. This was also confirmed in CRC, in which CiRS-7 promoted the proliferation and migration of CRC cells by regulating the EGFR/RAF1/mitogen-activated protein kinase (MAPK) pathway through inhibition of miR-7. Importantly, Kaplan–Meier analysis indicated that patients with high expression of CiRS-7 had significantly shorter OS than patients with low CiRS-7 expression did,76,77 implying that CiRS-7 is a promising prognostic biomarker in CRC patients and may serve as a therapeutic target for reducing EGFR/RAF1/MAPK activity in CRC patients.
CircHIPK3
CircHIPK3 is derived from exon 2 of homeodomain interacting protein kinase 3 (HIPK3).38 Recent studies have shown that circHIPK3 suppresses cancer cell proliferation and metastasis by sponging miR-182 and miR-558 in osteosarcoma78 and bladder cancer.79 However, its oncogenic role has been demonstrated in other cancers, such as hepatocellular carcinoma,80 ovarian cancer,81 glioma,82 nasopharyngeal carcinoma,83 lung cancer,84 and CRC.85 Zeng et al found that circHIPK3 was upregulated in CRC, and patients with high circHIPK3 expression had poorer OS than those with low circHIPK3 expression. Functionally, circHIPK3 can inhibit miR-7 activity, thereby leading to increased focal adhesion kinase (FAK), insulin-like growth factor 1 receptor (IGF1R), EGFR, and yin yang 1 (YY1) expression. In addition, luciferase reporter assays showed that the transcription factor c-Myb is an upstream regulator of circHIPK3 expression.85 These results suggest that circHIPK3 is a promising prognostic biomarker in CRC.
Circrna_100290
CircRNA_100290 was initially identified as a ceRNA that regulates CDK6 expression by sponging miR-29b in ovarian cancer.86 Subsequently, Fang et al reported that circRNA_100290 promoted the progression of CRC by regulating the expression of miR-516b and frizzled class receptor 4 (FZD4)-mediated activation of Wnt/β-catenin signaling. Furthermore, the expression of circRNA_100290 was positively correlated with tumor metastasis and inversely correlated with prognosis.87
CircCCDC66
CircCCDC66 is upregulated in all stages of colon cancer, and patients with higher levels of circCCDC66 have lower OS. Furthermore, the area under the receiver operating characteristic curve (AUC) calculated using the expression levels of circCCDC66 was 0.88, indicating that circCCDC66 is a promising predictive biomarker for CRC diagnosis and prognosis.88
Hsa_circ_0136666
Hsa_circ_0136666 is generated from the protein kinase, DNA-activated, catalytic subunit (PRKDC) gene, located on chr8:48715866–48730122.38 Hsa_circ_0136666 is overexpressed in CRC, and high expression levels were associated with poor OS of patients with CRC. Functional analysis with specifically designed siRNAs revealed that hsa_circ_0136666 regulates the proliferation and migration of CRC cells by sponging miR-136, thereby modulating the expression of SH2B adaptor protein 1 (SH2B1).89
Downregulated circRNAs in CRC
Hsa_circ_0001649
Hsa_circ_0001649 is produced from Snf2 histone linker PHD RING helicase (SHPRH), which serves as a tumor suppressor gene and a negative regulator of the Wnt/β-catenin signaling pathway.90 Hsa_circ_0001649 is reportedly downregulated in several types of cancers, including cholangiocarcinoma,91 gastric cancer,92 glioma,93,94 and hepatocellular carcinoma.95 Furthermore, hsa_circ_0001649 might serve as an independent prognostic factor for patients with glioma94 and hepatocellular carcinoma.95 Unlike circRNAs that serve as ceRNAs or miRNA sponges, hsa_circ_0001649 can produce a functional protein, named SHPRH-146aa, because it contains an open reading frame (ORF) driven by an internal ribosome entry site (IRES).93 Zhang et al found that hsa_circ_0001649 inhibited the proliferation and tumorigenicity of gliomas via SHPRH-146aa, which protected full-length SHPRH from degradation by ubiquitin-mediated proteolysis.93 In CRC, hsa_circ_0001649 was reported to be downregulated compared with the levels in non-tumorous tissues. The AUC of hsa_circ_0001649 was 0.857, suggesting that it could be used as a diagnostic biomarker in CRC.96 However, the functional mechanism of hsa_circ_0001649 in CRC requires further investigation.
CircITGA7
CircITGA7 is generated from exon 4 of integrin subunit alpha 7 (ITGA7) by back-splicing. Li et al found that circITGA7 was significantly downregulated in 91.38% of the CRC tissues (67/69) compared with expression in adjacent non-tumor tissues; the expression levels were inversely correlated with tumor size, lymph metastasis, distant metastasis, and TNM stage. The AUC of circITGA7 was 0.8791, with a sensitivity of 0.9275 and a specificity of 0.6667, indicating that circITGA7 has high diagnostic value for CRC. Functional studies revealed that circITGA7 binds to miR-370-3p and suppresses the Ras signaling pathway by upregulating neurofibromin 1 (NF1).97
Hsa_circ_0000711
Hsa_circ_0000711, a transcriptional product of the nuclear factor of activated T cells 3 (NFATC3) gene, was found to be significantly downregulated in CRC tissues. In addition, the AUC of hsa_circ_0000711 was 0.81, with a sensitivity of 0.91 and a specificity of 0.58. Moreover, Kaplan–Meier analysis showed that low expression of hsa_circ_0000711 could serve as an independent biomarker associated with poor OS of CRC patients.98
Hsa_circ_0014717
Derived from cyclin B1 (CCNB1), hsa_circ_0014717 was reported to be downregulated in gastric cancer13 and CRC.99 The decreased expression of hsa_circ_0014717 was notably associated with distant metastasis, TNM stage, and poor OS of CRC patients. Ectopic expression of hsa_circ_0014717 decreased the growth and invasion of CRC cells while inducing cell cycle G0/G1 phase arrest by upregulating p16 expression.99 Collectively, these results indicate that hsa_circ_0014717 has potential as a biomarker for the prognosis of CRC.
Hsa_circ_0000567
Hsa_circ_0000567, derived from exons 2–6 of SET domain-containing 3 (SETD3), was found to be significantly downregulated in CRC tissues compared with that in non-tumor tissues; this downregulation was closely correlated with tumor size, lymph metastasis, and distal metastasis in CRC. Importantly, the AUC of hsa_circ_0000567 was 0.87.100 These results indicate that hsa_circ_0000567 may represent a novel biomarker for the diagnosis of CRC and a novel target for treatment.
Hsa_circ_001988
Hsa_circ_001988, derived from FBXW7, was originally identified as a significantly downregulated circRNA in CRC by microarray, which was later verified by qRT-PCR. Among the assessed clinicopathological parameters, there was a significant association between the expression of hsa_circ_001988 and both differentiation and perineural invasion. Furthermore, the AUC of hsa_circ_001988 was 0.788, which indicates that hsa_circ_001988 could serve as a diagnostic biomarker.101
Hsa_circ_0026344
In a study of CRC tissues from 32 patients, hsa_circ_0026344 was shown to be significantly downregulated compared with the levels in paired adjacent non-tumorous tissues. The expression of hsa_circ_0026344 was correlated with tumor size and lymph metastasis as well as poor OS. Functionally, hsa_circ_0014717 overexpression significantly suppressed CRC cell proliferation and colony formation as well as promoted apoptosis by regulating miR-21 and miR-31 levels.102
Hsa_circ_0003906
The expression of hsa_circ_0003906 was examined in 122 matched CRC tissues and normal colon mucosa tissues. Compared with the levels in normal samples, hsa_circ_0003906 was significantly downregulated in the CRC samples. The results also demonstrated a significant correlation between the expression level of hsa_circ_0003906 and lymphatic metastasis and poor differentiation. In addition, the AUC of hsa_circ_0003906 for CRC was 0.818.103 Therefore, hsa_circ_0003906 may prove to be a useful diagnostic marker for patients with CRC.
Hsa_circ_104700
Based on circRNA arrays, Zhang et al identified 201 differentially expressed circRNAs, including 76 that were upregulated and 125 that were downregulated. Among them, hsa_circ_104700 was demonstrated to be downregulated in CRC tissues versus normal tissues. The expression of hsa_circ_104700 was significantly correlated with distal metastasis, and the AUC of hsa_circ_104700 was 0.616.104 These findings indicate that hsa_circ_104700 may serve as a biomarker for the diagnosis of CRC.
circRNAs in predicting distant metastasis
Zeng et al examined differentially expressed circRNAs in CRC tissues between patients with and without lung metastasis through microarray assays. A total of 431 circRNAs, including 192 upregulated and 239 downregulated circRNAs, were found to be differentially expressed in CRC tissues with lung metastasis compared with those without lung metastasis,105 revealing novel diagnostic biomarkers for CRC patients with lung metastasis. Xu et al detected circRNA expression in CRC tissues from three patients with and without liver metastasis through secondary sequencing. Ninety-two upregulated and 21 downregulated circRNAs were identified in CRC tissues with liver metastasis. Two of the circRNAs, hsa_circ_0001178 and hsa_circ_0000826, were found to be upregulated in 16 CRC tissues with liver metastasis compared with that in 24 CRC tissues without liver metastasis by qRT-PCR. The AUC was 0.945 for hsa_circ_0001178 and 0.816 for hsa_circ_0000826.106 These results indicate the potential diagnostic value of the two circRNAs for CRC patients with liver metastasis. Figure 1 summarizes the circRNAs involved in metastasis regulation.
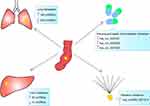 | Figure 1 CircRNAs that can be used to predict distant metastasis and response to chemoradiotherapy. CircRNA, circular RNA;↑, upregulated;↓, downregulated. |
circRNAs in predicting response to chemoradiotherapy
Researchers have found that the expression of circRNAs affects the sensitivity of tumors to chemotherapy and radiotherapy. For example, hsa_circRNA_0001313, also known as circCCDC66, has been reported to be upregulated in CRC tissues.88 Another study by Wang et al revealed that hsa_circRNA_0001313 was significantly upregulated in radioresistant CRC tissues compared with that in radiosensitive tissues. Knockdown of hsa_circRNA_0001313 reduced cell viability and colony formation and increased caspase-3 activity by negatively regulating miR-338-3p levels in CRC cells, thus inducing radiosensitivity.107 These results indicate that circ_0001313 is a potential biomarker of radiation resistance and a promising therapeutic target to overcome radiotherapy resistance in CRC. Using circRNA microarray analysis, Xiong et al identified 71 differentially expressed circRNAs between 5-fluorouracil (5FU)-based chemoradiation-resistant CRC cells and parental control cells. The upregulation of three of these circRNAs, hsa_circ_0007031, hsa_circ_0000504, and hsa_circ_0007006, was further confirmed in chemoradiation-resistant CRC cells by qRT-PCR, indicating their potential for prediction and prevention of chemoradiation resistance in CRC.108 Figure 1 summarizes the circRNAs that can be used to predict response to chemoradiotherapy.
Conclusions and perspectives
CircRNAs appear to be stably expressed in a tissue-specific manner and dysregulated in different cancers, including CRC. To date, numerous circRNAs have been found to be involved in various physiological and pathological processes in CRC, including proliferation, apoptosis, invasion, and migration. In addition to their most common function as miRNA sponges, some other functions, such as protein translation, transcriptional regulation, and cancer-associated signaling pathway regulation, have been revealed. Recently, the clinical applications of circRNAs in CRC have been explored by many investigators. As summarized in this review, a large number of circRNAs have been demonstrated as promising biomarkers for diagnosis and prognosis and predicting distant metastasis and response to chemoradiotherapy. Furthermore, synthetic circRNAs have been engineered for therapeutic purposes. An artificial circRNA was synthesized by Chang et al, and subsequent transfection into mammalian cells stimulated innate immunity gene expression and conferred protection against Venezuelan equine encephalitis viral infection.109 Accordingly, the transfection of synthetic circRNAs that produce specific therapeutic proteins into cancer cells is another research focus. The Anderson group has recently shown that artificial circular RNAs provide up to a threefold increase in the half-life of proteins and potent expression thereof in vitro, as compared with linear mRNA.110 Thus, we believe that synthetic circRNAs may be used for the treatment of CRC in the near future.
Nonetheless, circRNA research is still in its infancy, and multiple problems need to be resolved. First, a common problem is the lack of a unified standard for naming circRNAs. Second, circBase, which contains the most comprehensive information about circRNAs, still does not include many new circRNAs. Third, most researchers screen circRNA expression profiles using microarrays or RNA sequencing of a limited number of CRC tissues and adjacent normal tissues; therefore, errors in the results due to small sample size are likely. Importantly, we believe that the mechanism of circRNA participation in CRC development is complicated, and to date, limited functions have been identified. Therefore, more circRNAs await investigation, and their functions, mechanisms of action, and clinical applications require further elucidation.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Yuan W, Li X, Liu L, et al. Comprehensive analysis of lncRNA-associated ceRNA network in colorectal cancer. Biochem Biophys Res Commun. 2019;508(2):374–379. doi:10.1016/j.bbrc.2018.10.159
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
4. Wang X, Zhou J, Xu M, et al. A 15-lncRNA signature predicts survival and functions as a ceRNA in patients with colorectal cancer. Cancer Manag Res. 2018;10:5799–5806. doi:10.2147/CMAR.S178732
5. Pan S, Deng Y, Fu J, et al. TRIM52 promotes colorectal cancer cell proliferation throuth the STAT3 signaling. Cancer Cell Int. 2019;19:57. doi:10.1186/s12935-019-0775-4
6. Neve B, Jonckheere N, Vincent A, Van Seuningen I. Epigenetic regulation by lncRNAs: an overview focused on UCA1 in colorectal cancer. Cancers (Basel). 2018;10:11. doi:10.3390/cancers10110440
7. Liang Y, Zhang C, Ma MH, Dai DQ. Identification and prediction of novel non-coding and coding RNA-associated competing endogenous RNA networks in colorectal cancer. World J Gastroenterol. 2018;24(46):5259–5270. doi:10.3748/wjg.v24.i46.5259
8. Xu M, Chen X, Lin K, et al. lncRNA SNHG6 regulates EZH2 expression by sponging miR-26a/b and miR-214 in colorectal cancer. J Hematol Oncol. 2019;12(1):3. doi:10.1186/s13045-018-0690-5
9. Simon K. Colorectal cancer development and advances in screening. Clin Interv Aging. 2016;11:967–976. doi:10.2147/CIA.S109285
10. Greene J, Baird AM, Brady L, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. doi:10.3389/fmolb.2017.00038
11. Wang X, Fang L. Advances in circular RNAs and their roles in breast cancer. J Exp Clin Cancer Res. 2018;37(1):206. doi:10.1186/s13046-018-0870-8
12. Wang M, Yu F, Circular LP. RNAs: characteristics, function and clinical significance in hepatocellular carcinoma. Cancers (Basel). 2018;10:8. doi:10.3390/cancers10110400
13. Shao Y, Li J, Lu R, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6(6):1173–1180. doi:10.1002/cam4.1055
14. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
15. Yao R, Zou H, Liao W. Prospect of circular RNA in hepatocellular carcinoma: a novel potential biomarker and therapeutic target. Front Oncol. 2018;8:332. doi:10.3389/fonc.2018.00332
16. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19(2):141–157. doi:10.1261/rna.035667.112
17. Wang J, Zhu M, Pan J, Chen C, Xia S, Song Y. Circular RNAs: a rising star in respiratory diseases. Respir Res. 2019;20(1):3. doi:10.1186/s12931-019-1030-1
18. Qiu LP, Wu YH, Yu XF, Tang Q, Chen L, Chen KP. The emerging role of circular RNAs in hepatocellular carcinoma. J Cancer. 2018;9(9):1548–1559. doi:10.7150/jca.24566
19. Fu L, Jiang Z, Li T, Hu Y, Guo J. Circular RNAs in hepatocellular carcinoma: functions and implications. Cancer Med. 2018;7:3101–3109.
20. Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi:10.1016/j.molcel.2013.08.017
21. Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110(3). doi:10.1093/jnci/djx216.
22. Liu L, Liu FB, Huang M, et al. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat Dis Int. Epub 2019 Mar 9.
23. Pan H, Li T, Jiang Y, et al. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2018;119(1):440–446. doi:10.1002/jcb.27261
24. Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS One. 2015;10(6):e0131225. doi:10.1371/journal.pone.0131225
25. Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11(1):98. doi:10.1186/s13045-018-0643-z
26. Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. 2019;18(1):4. doi:10.1186/s12943-019-1010-6
27. Qin S, Zhao Y, Lim G, Lin H, Zhang X, Zhang X. Circular RNA PVT1 acts as a competing endogenous RNA for miR-497 in promoting non-small cell lung cancer progression. Biomed Pharmacother. 2018;111:244–250. doi:10.1016/j.biopha.2018.12.007
28. Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J Hematol Oncol. 2018;11(1):21. doi:10.1186/s13045-018-0569-5
29. Li T, Shao Y, Fu L, et al. Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl). 2018;96(1):85–96. doi:10.1007/s00109-017-1600-y
30. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi:10.1038/cr.2015.82
31. Chen S, Zhang L, Su Y, Zhang X. Screening potential biomarkers for colorectal cancer based on circular RNA chips. Oncol Rep. 2018;39(6):2499–2512.
32. Bachmayr-Heyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5:8057. doi:10.1038/srep08057
33. Zhang Z, Song N, Wang Y, et al. Analysis of differentially expressed circular RNAs for the identification of a coexpression RNA network and signature in colorectal cancer. J Cell Biochem. 2019;120(4):6409–6419. doi:10.1002/jcb.27928
34. Tian Y, Xu Y, Wang H, et al. Comprehensive analysis of microarray expression profiles of circRNAs and lncRNAs with associated co-expression networks in human colorectal cancer. Funct Integr Genomics. 2019;19(2):311–327. doi:10.1007/s10142-018-0641-9
35. Dou Y, Cha DJ, Franklin JL, et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982. doi:10.1038/srep37982
36. Jiang W, Zhang X, Chu Q, et al. The circular RNA profiles of colorectal tumor metastatic cells. Front Genet. 2018;9:34. doi:10.3389/fgene.2018.00173
37. He JH, Li YG, Han ZP, et al. The CircRNA-ACAP2/Hsa-miR-21-5p/Tiam1 regulatory feedback circuit affects the proliferation, migration, and invasion of colon cancer SW480 cells. Cell Physiol Biochem. 2018;49(4):1539–1550. doi:10.1159/000493457
38. Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi:10.1261/rna.043687.113
39. Yong W, Zhuoqi X, Baocheng W, Dongsheng Z, Chuan Z, Yueming S. Hsa_circ_0071589 promotes carcinogenesis via the miR-600/EZH2 axis in colorectal cancer. Biomed Pharmacother. 2018;102:1188–1194. doi:10.1016/j.biopha.2018.03.085
40. Li GF, Li L, Yao ZQ, Zhuang SJ. Hsa_circ_0007534/miR-761/ZIC5 regulatory loop modulates the proliferation and migration of glioma cells. Biochem Biophys Res Commun. 2018;499(4):765–771. doi:10.1016/j.bbrc.2018.03.219
41. Li B, Li X. Overexpression of hsa_circ_0007534 predicts unfavorable prognosis for osteosarcoma and regulates cell growth and apoptosis by affecting AKT/GSK-3β signaling pathway. Biomed Pharmacother. 2018;107:860–866. doi:10.1016/j.biopha.2018.08.086
42. Song L, Xiao Y. Downregulation of hsa_circ_0007534 suppresses breast cancer cell proliferation and invasion by targeting miR-593/MUC19 signal pathway. Biochem Biophys Res Commun. 2018;503(4):2603–2610. doi:10.1016/j.bbrc.2018.08.007
43. Qi Y, Zhang B, Wang J, Yao M. Upregulation of circular RNA hsa_circ_0007534 predicts unfavorable prognosis for NSCLC and exerts oncogenic properties in vitro and in vivo. Gene. 2018;676:79–85. doi:10.1016/j.gene.2018.07.028
44. Zhang R, Xu J, Zhao J, Wang X. Silencing of hsa_circ_0007534 suppresses proliferation and induces apoptosis in colorectal cancer cells. Eur Rev Med Pharmacol Sci. 2018;22(1):118–126.
45. Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691.
46. Shen H, Shen J, Wang L, et al. Low miR-145 expression level is associated with poor pathological differentiation and poor prognosis in non-small cell lung cancer. Biomed Pharmacother. 2015;69:301–305. doi:10.1016/j.biopha.2014.12.019
47. Xiong X, Sun D, Chai H, et al. MiR-145 functions as a tumor suppressor targeting NUAK1 in human intrahepatic cholangiocarcinoma. Biochem Biophys Res Commun. 2015;465(2):262–269. doi:10.1016/j.bbrc.2015.08.013
48. Schepeler T, Reinert JT, Ostenfeld MS, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68(15):6416–6424. doi:10.1158/0008-5472.CAN-07-6110
49. Andisheh-Tadbir A, Ashraf MJ, Jeiroodi N. Expression of CDK6 in oral squamous cell carcinomas. Asian Pac J Cancer Prev. 2018;19(4):1013–1016.
50. Xu XW, Zheng BA, Hu ZM, et al. Circular RNA hsa_circ_000984 promotes colon cancer growth and metastasis by sponging miR-106b. Oncotarget. 2017;8(53):91674–91683.
51. Zhang XL, Xu LL, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41(9):1056–1064.
52. Zhu Z, Tang J, Wang J, Duan G, Zhou L, Zhou X. MiR-138 acts as a tumor suppressor by targeting EZH2 and enhances cisplatin-induced apoptosis in osteosarcoma cells. PLoS One. 2016;11(3):e0150026. doi:10.1371/journal.pone.0150026
53. Zhu D, Gu L, Li Z, Jin W, Lu Q, Ren T. MiR-138-5p suppresses lung adenocarcinoma cell epithelial-mesenchymal transition, proliferation and metastasis by targeting ZEB2. Pathol Res Pract. 2019;215(5):28.
54. Jiang X, Wang J, Deng X, et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer. 2019;18:10. doi:10.1186/s12943-019-1010-6
55. Yang Z-G, Awan FM, Du WW, et al. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol Ther. 2017;25(9):2062–2074. doi:10.1016/j.ymthe.2016.10.004
56. Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. doi:10.1038/nrc2734
57. Du WW, Fang L, Yang W. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–370. doi:10.1038/cdd.2016.133
58. Xu Z, Li P, Fan L, Wu M. The potential role of circRNA in tumor immunity regulation and immunotherapy. Front Immunol. 2018;9:9. doi:10.3389/fimmu.2018.00009
59. Zhu M, Xu Y, Chen Y, Yan F. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138–144. doi:10.1016/j.biopha.2016.12.097
60. Han J, Zhao G, Ma X, et al. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Commun. 2018;503(4):2429–2435. doi:10.1016/j.bbrc.2018.06.172
61. Zhang J, Liu H, Zhao P, Zhou H, Mao T. Has_circ_0055625 from circRNA profile increases colon cancer cell growth by sponging miR-106b-5p. J Cell Biochem. 2019;120(3):3027–3037. doi:10.1002/jcb.27355
62. Guo JN, Li J, Zhu CL, et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451–7458. doi:10.2147/OTT.S123220
63. Wan L, Zhang L, Fan K, Cheng ZX, Sun QC, Wang JJ. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-catenin pathway. Biomed Res Int. 2016;2016:1579490. doi:10.1155/2016/1579490
64. Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6(8):6001–6013.
65. Jin Y, Yu LL, Zhang B, Liu CF, Chen Y. Circular RNA hsa_circ_0000523 regulates the proliferation and apoptosis of colorectal cancer cells as miRNA sponge. Braz J Med Biol Res. 2018;51(12):e7811. doi:10.1590/1414-431x20187811
66. Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–851. doi:10.1016/j.bbrc.2018.04.172
67. Bian L, Zhi X, Ma L, et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532-3p/FOXO4 axis. Biochem Biophys Res Commun. 2018;505(2):346–352. doi:10.1016/j.bbrc.2018.09.087
68. Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL, Yang Y. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res. 2018;37(1):325. doi:10.1186/s13046-018-1006-x
69. Wang Y, Mo Y, Gong Z, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16(1):25. doi:10.1186/s12943-017-0598-7
70. Zhang X, Yang D, Wei Y. Overexpressed CDR1as functions as an oncogene to promote the tumor progression via miR-7 in non-small-cell lung cancer. Onco Targets Ther. 2018;11:3979–3987. doi:10.2147/OTT.S158316
71. Su C, Han Y, Zhang H, et al. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-κB signalling. J Cell Mol Med. 2018;22(6):3097–3107. doi:10.1111/jcmm.13587
72. Li RC, Ke S, Meng FK, et al. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018;9(8):838. doi:10.1038/s41419-018-1111-y
73. Huang H, Wei L, Qin T, Yang N, Li Z, Xu Z. Circular RNA ciRS-7 triggers the migration and invasion of esophageal squamous cell carcinoma via miR-7/KLF4 and NF-κB signals. Cancer Biol Ther. 2019;20(1):73–80. doi:10.1080/15384047.2018.1507254
74. Zhang J, Hu H, Zhao Y, Zhao Y. CDR1as is overexpressed in laryngeal squamous cell carcinoma to promote the tumour’s progression via miR-7 signals. Cell Prolif. 2018;51(6):e12521. doi:10.1111/cpr.2018.51.issue-6
75. Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11(7):e0158347. doi:10.1371/journal.pone.0158347
76. Weng W, Wei Q, Toden S, et al. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2017;23(14):3918–3928. doi:10.1158/1078-0432.CCR-16-2541
77. Tang W, Ji M, He G, et al. Silencing CDR1as inhibits colorectal cancer progression through regulating microRNA-7. Onco Targets Ther. 2017;10:2045–2056. doi:10.2147/OTT.S131597
78. Xiao-Long M, Kun-Peng Z, Chun-Lin Z. Circular RNA circ_HIPK3 is down-regulated and suppresses cell proliferation, migration and invasion in osteosarcoma. J Cancer. 2018;9(10):1856–1862. doi:10.7150/jca.24619
79. Xie F, Li Y, Wang M, et al. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol Cancer. 2018;17(1):144. doi:10.1186/s12943-018-0892-z
80. Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9(2):175. doi:10.1038/s41419-018-1111-y
81. Liu N, Zhang J, Zhang LY, Wang L. CircHIPK 3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(12):3713–3718.
82. Jin P, Huang Y, Zhu P, Zou Y, Shao T, Wang O. CircRNA circHIPK3 serves as a prognostic marker to promote glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem Biophys Res Commun. 2018;503(3):1570–1574. doi:10.1016/j.bbrc.2018.07.081
83. Ke Z, Xie F, Zheng C, Chen D. CircHIPK 3 promotes proliferation and invasion in nasopharyngeal carcinoma by abrogating miR-4288-induced ELF3 inhibition. J Cell Physiol. 2019;234(2):1699–1706. doi:10.1002/jcp.27041
84. Yu H, Chen Y, Jiang P. Circular RNA HIPK3 exerts oncogenic properties through suppression of miR-124 in lung cancer. Biochem Biophys Res Commun. 2018;506(3):455–462. doi:10.1016/j.bbrc.2018.10.087
85. Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi:10.1038/s41419-018-1111-y
86. Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36(32):4551–4561. doi:10.1038/onc.2017.89
87. Fang G, Ye BL, Hu BR, Ruan XJ, Shi YX. CircRNA_100290 promotes colorectal cancer progression through miR-516b-induced downregulation of FZD4 expression and Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2018;504(1):184–189. doi:10.1016/j.bbrc.2018.08.152
88. Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77(9):2339–2350. doi:10.1158/0008-5472.CAN-16-1883
89. Jin C, Wang A, Liu L, Wang G, Li G. Hsa_circ_0136666 promotes the proliferation and invasion of colorectal cancer through miR-136/SH2B1 axis. J Cell Physiol. 2019;234(5):7247–7256. doi:10.1002/jcp.v234.5
90. Qu Y, Kalland KH, Ke X. Small molecule induces Wnt asymmetry in cancer. Cell Cycle. 2017;16(2):141–142. doi:10.1080/15384101.2016.1235850
91. Xu Y, Yao Y, Zhong X, et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496(2):455–461. doi:10.1016/j.bbrc.2018.01.077
92. Li WH, Song YC, Zhang H, et al. Decreased expression of Hsa_circ_00001649 in gastric cancer and its clinical significance. Dis Markers. 2017;2017:4587698. doi:10.1155/2017/4587698
93. Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–1814. doi:10.1038/s41388-017-0019-9
94. Wang Y, Sui X, Zhao H, et al. Decreased circular RNA hsa_circ_0001649 predicts unfavorable prognosis in glioma and exerts oncogenic properties in vitro and in vivo. Gene. 2018;676:117–122. doi:10.1016/j.gene.2018.07.037
95. Zhang X, Qiu S, Luo P, et al. Down-regulation of hsa_circ_0001649 in hepatocellular carcinoma predicts a poor prognosis. Cancer Biomark. 2018;22(1):135–142. doi:10.3233/CBM-171109
96. Ji W, Qiu C, Wang M, Mao N, Wu S, Dai Y. Hsa_circ_0001649: A circular RNA and potential novel biomarker for colorectal cancer. Biochem Biophys Res Commun. 2018;497(1):122–126. doi:10.1016/j.bbrc.2018.02.036
97. Li X, Wang J, Zhang C, et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J Pathol. 2018;246(2):166–179. doi:10.1002/path.5125
98. Li J, Ni S, Zhou C, Ye M. The expression profile and clinical application potential of hsa_circ_0000711 in colorectal cancer. Cancer Manag Res. 2018;10:2777–2784. doi:10.2147/CMAR.S172388
99. Wang F, Wang J, Cao X, Xu L, Chen L. Hsa_circ_0014717 is downregulated in colorectal cancer and inhibits tumor growth by promoting p16 expression. Biomed Pharmacother. 2018;98:775–782. doi:10.1016/j.biopha.2018.01.015
100. Wang J, Li X, Lu L, He L, Hu H, Circular XZ. RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. J Clin Lab Anal. 2018;32(5):e22379. doi:10.1002/jcla.22379
101. Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8(12):16020–16025.
102. Yuan Y, Liu W, Zhang Y, Zhang Y, Sun S. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem Biophys Res Commun. 2018;503(2):870–875. doi:10.1016/j.bbrc.2018.06.089
103. Zhuo F, Lin H, Chen Z, Huang Z, Hu J. The expression profile and clinical significance of circRNA0003906 in colorectal cancer. Onco Targets Ther. 2017;10:5187–5193. doi:10.2147/OTT.S147378
104. Zhang P, Zuo Z, Shang W, et al. Identification of differentially expressed circular RNAs in human colorectal cancer. Tumour Biol. 2017;39(3):1010428317694546.
105. Zeng Y, Xu Y, Shu R, et al. Altered expression profiles of circular RNA in colorectal cancer tissues from patients with lung metastasis. Int J Mol Med. 2017;40(6):1818–1828.
106. Xu H, Wang C, Song H, Xu Y, Ji G. RNA- Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers. Mol Cancer. 2019;18(1):8. doi:10.1186/s12943-019-1010-6
107. Wang L, Peng X, Lu X, Wei Q, Chen M, Liu L. Inhibition of hsa_circ_0001313 (circCCDC66) induction enhances the radio-sensitivity of colon cancer cells via tumor suppressor miR-338-3p: effects of cicr_0001313 on colon cancer radio-sensitivity. Pathol Res Pract. 2019;215(4):689–696. doi: 10.1016/j.prp.2018.12.032.
108. Xiong W, Ai YQ, Li YF, et al. Microarray analysis of circular RNA expression profile associated with 5-fluorouracil-based chemoradiation resistance in colorectal cancer cells. Biomed Res Int. 2017;2017:8421614. doi:10.1155/2017/8421614
109. Grace Chen Y, Kim MV, Chen X, et al. Sensing self and foreign circular RNAs by intron identity. Mol Cell. 2017;67(2):228–238. doi:10.1016/j.molcel.2017.05.022
110. Alexander Wesselhoeft R, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9:2629. doi:10.1038/s41467-018-05096-6
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
