Back to Journals » Infection and Drug Resistance » Volume 11
Emergence of tigecycline resistance in Escherichia coli co-producing MCR-1 and NDM-5 during tigecycline salvage treatment
Authors Wang Q, Zhang P, Zhao D , Jiang Y, Zhao F, Wang Y, Li X , Du X, Yu Y
Received 9 July 2018
Accepted for publication 21 September 2018
Published 13 November 2018 Volume 2018:11 Pages 2241—2248
DOI https://doi.org/10.2147/IDR.S179618
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Qian Wang,1 Ping Zhang,2,3 Dongdong Zhao,2 Yan Jiang,2,3 Feng Zhao,4 Yanfei Wang,3 Xi Li,5 Xiaoxing Du,2 Yunsong Yu2,3
1Department of General Practice, Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University, Hangzhou, China; 2Department of Infectious Diseases, Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University, Hangzhou, China; 3Key Laboratory of Microbial Technology and Bioinformatics of Zhejiang Province, Hangzhou, China; 4Department of Clinical Laboratory, Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University, Hangzhou, China; 5Centre of Laboratory Medicine, Zhejiang Provincial People’s Hospital, People’s Hospital of Hangzhou Medical College, Hangzhou, China
Objective: Here, we report a case of severe infection caused by Escherichia coli that harbored mcr-1, bla NDM-5, and acquired resistance to tigecycline during tigecycline salvage therapy.
Methods: Antimicrobial susceptibility testing, Southern blot hybridization, and complete genome sequence of the strains were carried out. The genetic characteristics of the mcr-1 and bla NDM-5 plasmids were analyzed. The whole genome sequencing of mcr-1-containing plasmid was completed. Finally, putative single nucleotide polymorphisms and deletion mutations in the tigecycline-resistant strain were predicted.
Results: Three E. coli isolates were obtained from ascites, pleural effusion, and stool of a patient; they were resistant to almost all the tested antibiotics. The first two strains separated from ascites (E-FQ) and hydrothorax (E-XS) were susceptible to amikacin and tigecycline; however, the third strain from stool (E-DB) was resistant to tigecycline after nearly 3 weeks’ treatment with tigecycline. All three isolates possessed both mcr-1 and bla NDM-5. The bla NDM-5 gene was found on the IncX3 plasmid, whereas the mcr-1, fosA3 and blaCTX-M-14 were located on the IncHI2 plasmid. Mutations in acrB and lon were the reason for the resistance to tigecycline.
Conclusion: This is the first report of a colistin-, carbapenem-, and tigecycline-resistant E. coli in China. Tigecycline resistance acquired during tigecycline therapy is of great concern for us because tigecycline is a drug of last resort to treat carbapenem-resistant Gram-negative bacterial infections. Furthermore, the transmission of such extensively drug-resistant isolates may pose a great threat to public health.
Keywords: mcr-1, blaNDM-5, tigecycline-resistance, acrB, lon
Introduction
The emergence of multidrug-resistant Gram-negative bacteria, especially those producing carbapenemases, continues to threaten public health because of the limited availability of antimicrobials for treatment. Colistin and tigecycline are drugs of last resorts to treat these severe infections. The rapid transmission of the colistin-resistance gene mcr-1 located on mobile elements has caused great concern since its initial discovery. Co-carriage of mcr-1 and blaNDM-5 has been reported in isolates from animals, food, and even humans since 2016.1–5 Tigecycline is effective against most carbapenemase-producing bacteria and was recently approved for clinical use in China; nevertheless, acquisition of resistance to tigecycline during therapy has been frequently reported.6–10 The mechanism of tigecycline resistance has not yet been clearly elucidated. It has been reported that resistance-nodulation-division (RND)-type transporters, primarily the AcrAB efflux pump, play an important role.11 Here, we report a case of intra-abdominal infection and pyothorax. Three extensively drug-resistant (XDR) Escherichia coli isolates with resistance to colistin and carbapenems were isolated from the patient. The first two strains, from ascites (E-FQ) and hydrothorax (E-XS), were susceptible to amikacin and tigecycline; however, the third strain from stool (E-DB) acquired resistance to tigecycline during tigecycline therapy. Fortunately, following combined antibiotic susceptibility tests in vitro, the infection was finally controlled.
To the best of our knowledge, this is the first report of a colistin-, carbapenem-, and tigecycline-resistant E. coli in China. The aim of our study was to clarify the resistance mechanisms of these XDR pathogens.
Materials and methods
Patient and isolate data
Three carbapenem-resistant strains of E. coli were isolated from a 46-year-old male patient. The patient had a history of hepatocellular carcinoma and was admitted to our hospital for intra-abdominal infection and pyothorax after liver resection in March 2017. To identify the bacteria and guide the antibiotic treatment, ascites and hydrothorax were cultured, and the patient underwent a stool screening. The first two strains cultured from ascites and hydrothorax were separated on March 8, 2017. The third, from stool, was isolated on March 28, 2017. All three isolates were identified by the VITEK 2 system (Sysmex-bioMérieux, Marcy l’Etoile, France).
PCR and Sanger sequencing
PCR and sequencing were used to identify the mcr-1 and blaNDM-5 genes in all three isolates. Mutations in acrB and lon were confirmed by PCR and Sanger sequencing (primers in Table S1).
Antimicrobial susceptibility test
Antibiotic susceptibilities, including imipenem, meropenem, amikacin, levofloxacin, cefotaxime, aztreonam, amoxicillin–clavulanic acid, and fosfomycin, were determined by the broth microdilution method according to the guidelines provided by the Clinical and Laboratory Standards Institute.12 Minimum inhibitory concentrations (MICs) of tigecycline and colistin were interpreted by EUCAST guidelines (http://www.eucast.org/). MIC by Etest followed the manufacturer’s instructions for some drugs, including cefoperazone/sulbactam, ciprofloxacin, aztreonam, and trimethoprim–sulfamethoxazole. E. coli ATCC 25922 was used as a quality control strain for the antibiotic susceptibility test. The interactions of amoxicillin–clavulanic acid and aztreonam were assessed using the checkerboard method13 with slight modifications. The combinations in the 96-well plates were performed as follows: aztreonam was diluted by twofold dilutions along the x-axis of the plates (from 1/64 MIC to 2 MIC), while amoxicillin–clavulanic acid was diluted by twofold dilutions along the y-axis (from 1/32 MIC to 2 MIC). Subsequently, each well was inoculated with the tested bacterial suspension (5×105 CFU/mL). The plates were then incubated at 37°C overnight. The interaction between aztreonam and amoxicillin–clavulanic acid was determined by quantifying the fractional inhibitory concentration index (FICI) using the following formula: FIC of aztreonam = MIC of aztreonam in combination/MIC of aztreonam alone; FIC of amoxicillin–clavulanic acid = MIC of amoxicillin–clavulanic acid in combination/MIC of amoxicillin–clavulanic acid alone. FICI = FICA + FICB. “Synergy” was defined when FICI ≤0.5; 0.5 < FICI ≤0.75 means “partial synergy”; 0.76 < FICI ≤1 denotes “additive”; 1 < FICI ≤4 denotes “indifferent”; while “antagonistic” in cases in which the FICI >4.
Pulsed-field gel electrophoresis (PFGE) and Southern blotting analysis
Genetic relationships among the isolates were evaluated by PFGE. Genomic DNA was digested with restriction enzyme XbaI (TaKaRa, Dalian, China) and was electrophoresed on a CHEF-mapper XA PFGE system (Bio-Rad, Berkeley, CA, USA) at 14°C and 6 V/cm and with alternating pulses at a 120° angle in a 5–35 seconds pulse time gradient for 22 hours. To ascertain the plasmid locations, genomic DNA was digested with S1-nuclease (TaKaRa, Kusatsu, Japan) and was electrophoresed for 18 hours at 14°C with run conditions of 6 V/cm and pulse times from 2.16 to 63.8 seconds. The DNA fragments were transferred to a positively charged nylon membrane (Millipore, Billerica, MA, USA) and then were hybridized with a digoxigenin-labeled blaNDM-5 and mcr-1-specific probe. An NBT/BCIP color detection kit (Roche, Mannheim, Germany) was then used to detect the target gene. The Salmonella enterica serotype Braenderup H9812 was used as the size marker.
Genomic DNA extraction and analysis
Genomic DNA from strain E-FQ, E-XS, E-DB was sequenced with an Illumina HiSeq 2000 (Illumina Inc., San Diego, CA, USA) following a paired-end 2×100-bp protocol. The total genomic DNA of E-DB was also extracted and sequenced via the single molecule real-time technique using a PacBio RS II platform and was assembled de novo using Canu v1.5. Gene prediction and annotation were performed with in silico online tools (http://rast.nmpdr.org/rast.cgi). Sequence comparison was performed using BLAST (http://blast.ncbi.nlm.nih.gov). Determination of plasmid replicons, resistance genes, and multi-locus sequence typing (MLST) was performed using online CGE tools (http://www.genomicepidemiology.org/). A comparison of genetic environment of mcr-1- and blaNDM-5-containing plasmids and their related plasmids was performed with EasyFig 2.2.2.14 The reads of E-DB were mapped against the reference genome of E-FQ and E-XS with the CLC Genomics Workbench 9 software (Qiagen, Valencia, CA, USA). The putative single nucleotide polymorphisms and deletion mutations were predicted.
Nucleotide sequence accession numbers
The next-generation sequencings of E-FQ, E-XS, and E-DB were subjected to National Center for Biotechnology Information under accession numbers PVQI00000000, PVQH00000000, and PVQG00000000. The complete sequences of pE-DB-mcr were submitted to GenBank under accession number MH128771.
Results
Patient information
A 46-year-old male patient presented to our hospital (a tertiary-care hospital in Hangzhou) with recurrent fever, pyothorax, and intra-abdominal infection in March 2017. The patient had a history of chronic hepatitis B and liver cancer. He had undergone a liver resection for the hepatocellular cancer 9 months prior. The patient had recurrent fever and abdominal pain after the surgery. A colonic fistula was found on imaging studies and during enteroscopy. The patient was treated with imipenem (0.5 g q8h for 8 days) and tigecycline (50 mg q12h for 7 days) prior to this admission.
The results of laboratory examinations were as follows: white blood cell count, 15,200/mm3; neutrophil percentage, 91%; platelet count, 79,000/mm3; alpha-fetoprotein, 1,238.3 ng/mL; C-reactive protein, 62.6 mg/L, and procalcitonin, 0.5 µg/L. A series of pathogen cultures from ascites, plural effusion, and stool grew XDR E. coli that were resistant to almost all tested antibiotics. The first two strains separated from ascites (E-FQ) and hydrothorax (E-XS) after admission were susceptible to amikacin and tigecycline; however, the third strain from stool (E-DB) was susceptible to amikacin but was resistant to tigecycline after nearly 3 weeks’ treatment with tigecycline (50 mg q12 h for 17 days and 100 mg q12 h for 3 days) in our hospital.
The patient underwent ileum colostomy and abdominal cavity drainage, while amoxicillin–clavulanic acid combined with aztreonam was used for 1 week according to the in vitro combined antibiotic susceptibility test (FICI 0.125). The bacterium was finally eliminated. However, the patient died in the fourth month of hospitalization in our hospital because of cancer recurrence.
Isolate characteristics
The antimicrobial susceptibility testing results showed that three E. coli strains were resistant to almost all tested antibiotics (Table 1). The first two strains E-FQ and E-XS were susceptible to amikacin (MIC =8 mg/L) and tigecycline (MIC =1 mg/L); however, the third strain E-DB was resistant to tigecycline (MIC =8 mg/L). PCR and sequencing identified that all three isolates contained genes mcr-1 and blaNDM-5. These isolates expressed indistinguishable band patterns by XbaI PFGE analysis (Figure S1). In addition to mcr-1 and blaNDM-5, the three strains were found to harbor resistance genes for aminoglycosides (aph(4)-Ia, aadA1, strB, strA, aacA4, aac(3)-IId and aac(3)-IVa), beta-lactam (blaCTX-M-14), fluoroquinolone (aac(6’)Ib-cr), fosfomycin (fosA), phenicol (floR), sulfonamide (sul1 and sul2), trimethoprim (dfrA1), and tetracycline (tet(A), that were consistent with an extremely drug-resistant profile. Plasmid incompatibility typing detected the presence of the plasmid replicons IncA/C2, IncFIA, IncHI2A, IncHI2, IncX3, and IncFIB. MLST demonstrated that the isolates belonged to sequence type 2736. The reads of strain E-DB were mapped against the reference genome of E-FQ and E-XS. The difference between E-DB and E-FQ was less than five single nucleotide variants (Table 2), meanwhile, the difference between E-DB and S-XS was also less than five.
  | Table 2 The putative single nucleotide variants and deletion mutations in Escherichia coli E-DB compared with E-FQ and E-XS Abbreviation: ORF, open reading frame. |
Genetic context of plasmids
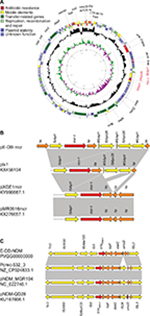
S1 nuclease PFGE and Southern blot showed the presence of mcr-1 in an ~244–330 kb plasmid and blaNDM-5 in an ~33–55 kb plasmid (Figure S1), belonging to IncHI2 and IncX3 by analysis of whole genome sequencing. mcr-1-containing plasmid pE-DB-mcr was 230,365 bp in size with an average G+C content of 51%. In the multidrug resistance (MDR) region, it harbored resistance genes for aminoglycosides (aph(4)-Ia, aacA4, and aac(3)-IVa), fosfomycin (fosA3), phenicol (floR), sulfonamide (sul2) and beta-lactam (blaCTX-M-14). Around the colistin resistance gene mcr-1, it had a structure of ISApl1-mcr-1-hp-#ISApl1(237 bp)-ISKpn26-#ISApl1(237 bp)-ISKpn26-#ISApl1 (837 bp) (Figure 1A). The mcr-1 element was flanked by two ISApl1 elements in the same orientation, and the ISApl1 upstream was inserted by an ISKpn26 into two parts (837 bp and 237 bp), with a duplication of ISKpn26 and one part of ISApl1 (237 bp) that was highly similar to plasmid pls1 (GenBank accession no. KX458104) (Figure 1B). BLASTn comparisons demonstrated that the genetic surrounding of blaNDM-5 was relatively similar to the sequences of the blaNDM-5-carrying plasmid pCREC-532_3 (GenBank accession no. NZ_CP024833.1), pNDM_MGR194 (GenBank accession no. NC_022740.1), and pNDM-QD28 (GenBank accession no. KU167608.1), with a structure of Tn3-IS3000-ISAba125-IS5-blaNDM-5-trpF-dsbC-ISL3 (Figure 1C).
Tigecycline resistance
Antimicrobial susceptibility showed that E-FQ and E-XS were sensitive to tigecycline (MIC 1 mg/L), while strain E-DB was resistant to tigecycline (MIC 8 mg/L). The putative single nucleotide and deletion mutations in E-DB were predicted. acrB and lon were the only two genes found in both mappings (Table 2). Mutations in acrB and lon were confirmed by PCR and Sanger sequencing. As to acrB, it is a single nucleotide mutation that causes a change of arg to cys (R620C). As to lon, it is a single nucleotide polymorphism mutation that resulted in the change of pro to leu (P403L) (Table 2).
Discussion
To our knowledge, this is the first report of an isolate harboring mcr-1 and blaNDM-5 acquiring resistance to tigecycline during therapy. Since its discovery in Southern China, mcr-1 has already been discovered in >40 countries/regions covering five of seven continents and where it plays an important role in polymyxin resistance.15 Searches for mcr-1-bearing plasmids deposited in GenBank suggest that IncHI2 is one of the most dominant replicon types of plasmid carrying mcr-1 plasmids (45/219, 20.5%); IncHI2 plasmids had the largest size, with sequence lengths up to 267,486 bp that were always integrated alongside a large MDR gene cassette.16 In our study, we found that the mcr-1-containing plasmid also harbored the fosfomycin resistance gene fosA3 and beta-lactam blaCTX-M-14. The horizontal diffusion of these resistance genes is particularly worrisome because the regimen of colistin plus fosfomycin remains a therapeutic alternative in cases of multidrug-resistant bacteria. There is increasing evidence that ISApl1 plays a pivotal role in the mobilization of mcr-1. Tn6330, with the structure ISApl1-mcr-1-orf-ISApl1, was proposed as a key element mediating translocation of mcr-1 into various plasmid backbones through formation of a circular intermediate.17,18 In our study, the ISApl1 in upstream of Tn6330 was inserted by an ISKpn26 in two parts, and we infer that this insertion may increase the stability of Tn6330 in the backbone of IncHI2.
Tigecycline belongs to the group called glycylcyclines that binds to 16S rRNA and prevents mRNA decoding. Compared with earlier class tetracyclines, the bulky side chain attached to the C-9 position of ring D is the key to escape common tetracycline resistance mechanisms, for example, tetracycline efflux pumps and ribosomal protection.19 Tigecycline is also not affected by most of the common mechanisms that affect other classes of antimicrobial agents, including target site modifications, enzymatic degradation of the drug molecule, and DNA gyrase mutations.20 However, the reports of clinical tigecycline resistance were published soon after its use in medical practice. Tigecycline resistance occurring in carbapenem-resistant Enterobacteriaceae (CRE) during therapy has been reported in many cases.7–10 It had been reported that Tet proteins can mutate to acquire high-level tigecycline resistance,21 but in most cases, tigecycline resistance is due to the constitutive expression of RND efflux pumps; for example, AcrAB in E. coli.22 In E. coli, the tripartite efflux system AcrAB-TolC is the pump in charge of the efflux of dyes, bile salts, detergents, and a large group of antibiotics, including the tetracyclines. AcrB is an RND pump situated in the inner membrane that functions in a complex with two other proteins, an outer membrane channel TolC and a periplasmic adaptor protein AcrA. AcrB is the major site for substrate recognition and energy transduction of the entire tripartite system.23,24 In one study, the author found a trend toward higher acrB expression as tigecycline MIC increases.25 Several global transcriptional regulators of the araC family, ramA, marA, soxS, and rarA, may participate in tigecycline resistance via AcrAB efflux pump activation,25–27 of which ramA, marA, and soxS are regulated by the lon.28,29 Nicoloff et al found that about half of the selected spontaneous tetracycline- or chloramphenicol-resistant E. coli mutants carried additional lon mutations.30 lon mutation causes low MDR by stabilizing MarA and SoxS, two transcriptional activators, whose induction increases resistance to many antimicrobial drugs. The first-step lon mutation greatly increases the rate of development of higher resistance via acquisition of new sets of mutations.29 In this case, with tigecycline treatment, mutations in acrB (R620C) and lon (P403L) in strain E-DB caused activation of the AcrAB-TolC efflux pump, leading to the high MIC of 8 mg/L for tigecycline.
CRE are often associated with severe health care-associated infections with high mortality ranging from 32.1% to 71%.31 NDM-type beta-lactamase are the second most common carbapenemases found among CRE in China.32 These carbapenemases cannot be inhibited by β-lactamase inhibitors but remain susceptible to aztreonam, although most metallo-beta-lactamase-producing CRE isolates also harbor extended-spectrum β-lactamases that confer resistance to aztreonam. Therefore, many NDM producers remain susceptible only to tigecycline, colistin, and to a lesser extent to fosfomycin.33 In our disseminated infection case, the colistin- and tigecycline-resistant CRE made the treatment more complex and challenging, and even more worrisome, because the E. coli harbored fosA3 and blaCTX-M-14. In addition to the surgical intervention, amoxicillin–clavulanic acid combined with aztreonam was used on the basis of the combined antibiotic susceptibility test, and the bacterium was finally eliminated. This regimen may promisingly combat E. coli harboring mcr-1 and blaNDM-5. Combined antibiotic susceptibility test in vitro is important in the so-called “post-antibiotic” era.
Conclusion
This is the first report of a colistin-, carbapenem- and tigecycline-resistant E. coli in China. Tigecycline resistance acquired during tigecycline therapy is a great concern because tigecycline is a drug of the last resort for carbapenem-resistant Gram-negative bacteria infections. Furthermore, the transmission of such extensively drug-resistant isolates may pose a great threat to public health. Appropriate treatment schemes that effectively kill the XDR pathogen are imperative.
Ethics statement
The clinical isolates of E. coli E-FQ, E-XS, E-DB were part of routine hospital laboratory procedure. The patient’s next of kin had provided written informed consent for details to be used in the manuscript.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 81471986, No. 81772234, No. 31700125), the Medical and Health Technology Project of Zhejiang Province (2018RC011), and the Natural Science Young Foundation of Zhejiang Province, China (LQ17H190006).
Author contributions
Conceived and designed the experiments: YY and PZ; performed the experiments: QW, PZ, YW, and DZ; analyzed the data: DZ, FZ, and YJ; wrote the manuscript: QW, PZ, XL, and XD. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Yang RS, Feng Y, Lv XY, et al. Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single Muscovy duck (Cairina moschata). Antimicrob Agents Chemother. 2016;60(11):6899–6902. | ||
Yao X, Doi Y, Zeng L, Lv L, Liu JH. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis. 2016;16(3):288–289. | ||
Lai CC, Chuang YC, Chen CC, Tang HJ. Coexistence of MCR-1 and NDM-9 in a clinical carbapenem-resistant Escherichia coli isolate. Int J Antimicrob Agents. 2017;249(104):517–518. | ||
Zheng B, Lv T, Xu H, et al. Discovery and characterisation of an Escherichia coli ST206 strain producing NDM-5 and MCR-1 from a patient with acute diarrhoea in China. Int J Antimicrob Agents. 2018;51(2):273–275. | ||
Mediavilla JR, Patrawalla A, Chen L, et al. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. mBio. 2016;7(4):e01191–16. | ||
Niebel M, Quick J, Prieto AM, et al. Deletions in a ribosomal protein-coding gene are associated with tigecycline resistance in Enterococcus faecium. Int J Antimicrob Agents. 2015;46(5):572–575. | ||
Rodríguez-Avial C, Rodríguez-Avial I, Merino P, Picazo JJ. Klebsiella pneumoniae: development of a mixed population of carbapenem and tigecycline resistance during antimicrobial therapy in a kidney transplant patient. Clin Microbiol Infect. 2012;18(1):61–66. | ||
Spanu T, De Angelis G, Cipriani M, et al. In vivo emergence of tigecycline resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli. Antimicrob Agents Chemother. 2012;56(8):4516–4518. | ||
Tsai HY, Liao CH, Cheng A, et al. Emergence of tigecycline-resistant Klebsiella pneumoniae after tigecycline therapy for complicated urinary tract infection caused by carbapenem-resistant Escherichia coli. J Infect. 2012;65(6):584–586. | ||
van Duin D, Cober ED, Richter SS, et al. Tigecycline therapy for carbapenem-resistant Klebsiella pneumoniae (CRKP) bacteriuria leads to tigecycline resistance. Clin Microbiol Infect. 2014;20(12):O1117–O1120. | ||
Pournaras S, Koumaki V, Spanakis N, Gennimata V, Tsakris A. Current perspectives on tigecycline resistance in Enterobacteriaceae: susceptibility testing issues and mechanisms of resistance. Int J Antimicrob Agents. 2016;48(1):11–18. | ||
CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. (CLSI Supplement M100). Available from: clsi 2017 m100s 27th.pdf. Accessed October 25, 2018. | ||
Hornsey M, Wareham DW. In vivo efficacy of glycopeptide-colistin combination therapies in a Galleria mellonella model of Acinetobacter baumannii infection. Antimicrob Agents Chemother. 2011;55(7):3534–3537. | ||
Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010. | ||
Sun J, Zhang H, Liu YH, Feng Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. 2018;26(9):794–808. | ||
Matamoros S, van Hattem JM, Arcilla MS, et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Rep. 2017;7(1):15364. | ||
Snesrud E, He S, Chandler M, et al. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob Agents Chemother. 2016;60(11):6973–6976. | ||
Li R, Xie M, Zhang J, et al. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother. 2017;72(2):393–401. | ||
Sum PE, Petersen P. Synthesis and structure-activity relationship of novel glycylcycline derivatives leading to the discovery of GAR-936. Bioorg Med Chem Lett. 1999;9(10):1459–1462. | ||
Peterson LR. A review of tigecycline – the first glycylcycline. Int J Antimicrob Agents. 2008;32(Suppl 4):S215–S222. | ||
Linkevicius M, Sandegren L, Andersson DI. Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrob Agents Chemother. 2016;60(2):789–796. | ||
Linkevicius M, Sandegren L, Andersson DI. Mechanisms and fitness costs of tigecycline resistance in Escherichia coli. J Antimicrob Chemother. 2013;68(12):2809–2819. | ||
Pos KM. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta. 2009;1794(5):782–793. | ||
Blair JM, Piddock LJ. Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr Opin Microbiol. 2009;12(5):512–519. | ||
He F, Fu Y, Chen Q, et al. Tigecycline susceptibility and the role of efflux pumps in tigecycline resistance in KPC-producing Klebsiella pneumoniae. PLoS One. 2015;10(3):e0119064. | ||
Grkovic S, Brown MH, Skurray RA. Regulation of bacterial drug export systems. Microbiol Mol Biol Rev. 2002;66(4):671–701. | ||
Bratu S, Landman D, George A, Salvani J, Quale J. Correlation of the expression of acrB and the regulatory genes marA, soxS and ramA with antimicrobial resistance in clinical isolates of Klebsiella pneumoniae endemic to New York City. J Antimicrob Chemother. 2009;64(2):278–283. | ||
Ricci V, Blair JM, Piddock LJ. RamA, which controls expression of the MDR efflux pump AcrAB-TolC, is regulated by the Lon protease. J Antimicrob Chemother. 2014;69(3):643–650. | ||
Nicoloff H, Andersson DI. Lon protease inactivation, or translocation of the lon gene, potentiate bacterial evolution to antibiotic resistance. Mol Microbiol. 2013;90(6):1233–1248. | ||
Nicoloff H, Perreten V, Levy SB. Increased genome instability in Escherichia coli lon mutants: relation to emergence of multiple-antibiotic-resistant (Mar) mutants caused by insertion sequence elements and large tandem genomic amplifications. Antimicrob Agents Chemother. 2007;51(4):1293–1303. | ||
Livorsi DJ, Chorazy ML, Schweizer ML, et al. A systematic review of the epidemiology of carbapenem-resistant Enterobacteriaceae in the United States. Antimicrob Resist Infect Control. 2018;7:55. | ||
Zhang Y, Wang Q, Yin Y, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from the China CRE Network. Antimicrob Agents Chemother. 2018;62(2):e01882–17. | ||
Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18(5):263–272. |
Supplementary materials
  | Table S1 Primers used in mapping the mcr-1 genetic environment and confirming mutations in acrB and lon |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.