Back to Journals » Infection and Drug Resistance » Volume 16
Emergence of OXA-484-Producing Klebsiella variicola in China
Authors Ge H, Qiao J, Xu H, Liu R, Zhao J, Chen R, Li C , Chen M, Guo X
Received 20 January 2023
Accepted for publication 23 March 2023
Published 27 March 2023 Volume 2023:16 Pages 1767—1775
DOI https://doi.org/10.2147/IDR.S404551
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Haoyu Ge,1,2,* Jie Qiao,1,2,* Hao Xu,2 Ruishan Liu,2 Junhui Zhao,3 Ruyan Chen,1 Chenyu Li,1 Mantao Chen,4 Xiaobing Guo1
1Department of Laboratory Medicine, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, People’s Republic of China; 2Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, the First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, People’s Republic of China; 3School of Basic Medical Sciences, Zhejiang Chinese Medical University, Hangzhou, People’s Republic of China; 4Department of Neurosurgery, the First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaobing Guo, Department of Laboratory Medicine, The First Affiliated Hospital of Zhengzhou University, Jianshe East Road, Zhengzhou, Henan Province, 450000, People’s Republic of China, Tel +86 371 6627 8237, Fax +86 371 6691 3569, Email [email protected]
Purpose: The frequent and inappropriate use of antibiotics has caused a dramatic rise in the number, species, and degree of multi-drug resistant bacteria, making them more prevalent and difficult to treat. In this context, the aim of the present study was to characterize the OXA-484-producing strains isolated from a perianal swab of a patient by using whole-genome analysis.
Patients and Methods: In this study, carbapenemase-producing Klebsiella variicola was identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), average nucleotide identity (ANI) and PCR. S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and Southern blotting were utilized to characterize the plasmid profiles of K. variicola 4717. In particular, WGS was performed to obtain genomic information on this clinical isolate, and assemble all the plasmids of the blaOXA-484-harboring strain.
Results: The antimicrobial susceptibility pattern of K. variicola 4717 revealed that it was resistant to a range of antibiotics, including aztreonam, imipenem, meropenem, ceftriaxone, cefotaxime, ceftazidime, levofloxacin, ciprofloxacin, piperacillin-tazobactam, methylene-sulfamer oxazole, amoxicillin-clavulanic acid, cefepime, and tigecycline. Its susceptibility to chloromycin was intermediate, while it was still susceptible to amikacin, gentamicin, fosfomycin, and polymyxin B. The presence of two companion plasmids, p4717_1 and p4717_2, together with a plasmid carrying the blaOXA-484 gene was observed. An in-depth investigation of p4717-OXA-484 uncovered that it is an IncX3-type plasmid and shares a similar segment encoded by IS26. Given the similar genetic background, it was conceivable that blaOXA-484 could have developed from blaOXA-181 through a series of mutations.
Conclusion: Herein, we described the first genome sequence of K. variicola strain harbouring the class D β-actamase blaOXA-484 in an Inc-X3-type plasmid. Our work also uncovered the genetic characterization of K. variicola 4717 and the importance of initiating antimicrobial detection promptly.
Keywords: Klebsiella variicola, OXA-484, IncX3, mutation
Introduction
Klebsiella variicola, a species of the Klebsiella genus, exhibits genetic and biochemical distinctions in comparison to Klebsiella pneumoniae.1 K. variicola was first reported in 2004 and mainly isolated from natural environments.2 Additionally, they have an ability to fix nitrogen and promote the plant growth.3 As regarding its characteristic of antimicrobial resistance in recent years, K. variicola was considered as an emerging human pathogen which should be monitored.4 A hypermucoviscous multidrug-resistant K. variicola coproducing IMP-4 and NDM-1 was obtained from a pediatric patient.5 Multidrug-resistant K. variicola has caused outbreaks in a Bangladeshi neonatal unit with high mortality. Given all that, more attention should be paid to K. variicola, which may be a reservoir of antimicrobial resistance genes. However, in most clinical laboratories, K. variicola was often misidentified as K. pneumoniae, until the PCR and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) were used for species identification.6 In addition, K. variicola can also be recognized on the basis of average nucleotide identity (ANI). Thus, resistance mechanism and genetic characteristic of K. variicola remain largely unknown until now.
OXA-48 is a class of carbapenemases that mainly appear in European countries, Middle East and Mediterranean countries.7 To the best of our knowledge, at least 35 variants of OXA-48 have been reported, the difference between them being a few amino acid substitutions.8 Among them, OXA-484, which differs from OXA-48 in five amino acid substitutions (Thr104Ala, Asn110Asp, Glu168Gln, Ser171Ala, Arg214Gly), was collected from K. pneumoniae strains in the UK.9 It has also been documented that Escherichia coli is hosting the blaOXA-484 gene.10 In this study, we were the first to document the infection caused by OXA-484-producing K. variicola in China.
Materials and Methods
Species Identification and Antimicrobial Susceptibility Testing
Isolates were collected from a tertiary hospital in Zhengzhou, Henan province, China, during our routine surveillance of multi-drug resistance bacteria. The species was identified using both MALDI-TOF/MS (Bruker Daltonik GmbH, Bremen, Germany) and average nucleotide identity (ANI) analysis.11 The carbapenemase-encoding genes were detected using PCR and sanger sequencing. Antimicrobial susceptibility testing (AST) was conducted using the agar dilution method and broth microdilution method.12 The Clinical Laboratory Standards Institute (CLSI) guidelines (https://clsi.org/standards/) were used for the interpretation of susceptibility results. The resistance breakpoints from the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org) were used for polymyxin B and tigecycline. K. pneumoniae ATCC700603 and E. coli ATCC 25922 were used as quality controls strains.13
Localization and Transferability of blaOXA-484
The size and number of plasmids carried by K. variicola 4717 was visualized by S1-PFGE and southern blotting. The conjugation assay aimed to determine the transfer capacity of the blaOXA-484-harbouring plasmid. The sodium azide-resistant strain E. coli J53 used as the recipient and the blaOXA-484-positive K. variicola 4717 was selected as the donor. MH agar plates supplemented with NaN3 (200 mg/L) and imipenem (2 mg/L) as the screen plate for the blaOXA-484-positive transconjugants. Then, MALDI-TOF MS and PCR were used to confirm the presumptive conjugant.14
Whole Genome Sequencing and Bioinformatics Analysis
Genomic DNA of K. variicola 4717 was obtained through a Bacterial DNA Kit (QIAGEN, Hilden, Germany) according to the instruction.15 The extracted genome of K. variicola 4717 was sequencing on the Illumina NovaSeq 6000 (Illumina, San Diego, CA, United States) for short-read data and Oxford Nanopore platform (Oxford Nanopore Technologies, Oxford, United Kingdom) for long-read data at different depths.16 Then, the Illumina short reads and Nanopore long reads were assembled using Unicycler for whole genome sequence. Prokka (https://www.psc.edu/resources/software/prokka/) was used for genome annotation. Genomic data were analyzed using various online tools. ResFinder (https://cge.food.dtu.dk/services/ResFinder/) and the BacWGSTdb (http://www.bacdb.cn/BacWGSTdb/) were employed to detect acquired antimicrobial resistance genes and virulence genes, respectively.17 Multilocus sequence typing (MLST) of K. variicola isolates by analyzing the seven housekeeping genes (leuS, pgi, pgk, phoE, pyrG, rpoB, and fusA) using an online database (http://mlstkv.insp.mx/).18 Plasmid finder was used to identify the replicon type of plasmids (https://cge.food.dtu.dk/services/PlasmidFinder/). The origin of transfers in DNA sequences of bacterial mobile genetic elements was identified on oriTfinder (https://tool-mml.sjtu.edu.cn/oriTfinder/oriTfinder.html).19 The NCBI blast was used to align the plasmids which were similar to the blaOXA-484-harbouring plasmid. The comparisons images of multiple plasmids were conducted by using the BLAST Ring Image Generator (BRIG) in concentric rings and the figures of genetic context surrounding the blaOXA-484-harbouring plasmid and other related plasmids were generated by Easyfig 2.0 software in linear graph.
Biofilm Formation Assays
Biofilm formation was measured in accordance with the assays outlined in previous researches.20 In a word, the bacterial overnight culture was diluted in LB and the 200 μL of the mixture was dispensed into the each well of 96-well plate. After static culture at 37°C for 24 hours, PBS was utilized to clean the microwells three times in order to eliminate all non-adherent bacteria. Methanol was used for fixation, and 0.1% crystal violet solution were added to each well for subsequent stain. After washing the plate with PBS three times and discarding the washing solution, 100 μL of DMSO were added to dissolve the crystal violet attached to the biofilm, and the plate should be incubated for 5 minutes. Finally, the OD (optical density) was measured at 590 nm. LB broth was used for the negative control.
Results
Species Identification
The isolate FAHZZU 4717 was obtained from a stored strain which was collected from a perianal swab of a patient admitted to ICU in the First Affiliated Hospital of Zhengzhou University in 2021. FAHZZU 4717 was identified as K. variicola based on the ANI analysis. Moreover, the genomic sequences of FAHZZU 4717 share 99.24% similarity and 87% coverage with the genome of the variicola reference strain F2R9, which has been archived as ATCC BAA-830 (no. CP072130).
Antimicrobial Susceptibility Profiles of K. variicola 4717
K. variicola 4717 showed intermediate to chloromycin, displayed susceptibility to amikacin, gentamicin, fosfomycin, tigecycline, polymyxin B, and showed resistance to aztreonam, imipenem, meropenem, ceftriaxone, cefotaxime, ceftazidime, levofloxacin, ciprofloxacin, piperacillin-tazobactam, methylene-sulfamer oxazole, amoxicillin-clavulanic acid, and cefepime (Table 1).
 |
Table 1 Susceptibility of K. variicola 4717 |
Characterization of the Genome of K. variicola 4717
The genome of K. variicola 4717 consists of six contigs. Among them, contig 1 (5,672,190 bp) belong to the chromosome, while the contig 2 (168,009 bp), contig 3 (115,032 bp) and contig 4 (51,479) belong to three different plasmids (Table S1). However, only two plasmid replicons were identified, contig 2 belongs to the IncFII and IncFIB(K), and contig 4 belongs to the IncX3 which designated p4717-OXA-484. Moreover, MLST analysis showed that the allelic profile (leuS, pgi, pgk, phoE, pyrG, rpoB, and fusA) of K. variicola 4717 was 25:24:6:1:34:1:4, which indicates an unknown ST.
The ARGs existing in the genome of K. variicola 4717 were presented in Table S1. We identified the quinolone resistance genes oqxB, oqxA, and qnrS1; beta-lactam resistance genes blaLEN16, blaDHA-1, and blaOXA-484; fosfomycin resistance gene fosA; sulphonamide resistance gene sul1; trimethoprim resistance gene dfrA1, and tetracycline resistance gene tet(A).
K. variicola 4717 was found to possess approximately 60 virulence factors (Table S2), including those related to adhesion (fimH, fimF, and fimC), type VI secretion systems (vipA/tssB, impA/tssA, dotU/tssL, vasE/tssK, hcp/tssD, and sciN/tssJ), iron acquisition (entA, entD, entE, fepA, fepB, and fes), and biofilm formation (ompA, mrkC, mrkD, and mrkH). In addition, experiments on biofilm formation demonstrated that K. variicola 4717 has a moderate capacity for biofilm production (Figure 1).
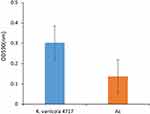 |
Figure 1 Biofilm formation analysis of K. variicola 4717. Ac, the negative control. |
Characterization of blaOXA-484 Bearing Plasmid
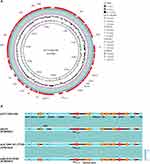
The p4717-OXA-484 is 51,457 bp long, with an average GC content of 46.4% (Figure S1). Its replicon was identified as the IncX3 incompatibility group. Combined with the result of BLASTn search, linear alignment of the plasmid genomes suggests that p4717-OXA-484 is similar to three different plasmids from E. coli, K. pneumoniae, and K. variicola, namely pLB_OXA-181_PT109 (no. CP041033), pSECR18-2374D (no. CP041931), and pKS22 (no. KT005457), respectively (Figure 2A). Additionally, like the three plasmids mentioned above, the blaOXA-484 was consistently associated with a composite transposon which was flanked by two copies of IS26. The similar region contains transposons (Tn2), insertion sequences (ISKpn19, IS3000) and other genes (tnpR, umuD, DNAase and qnrS1) (Figure 2B). There were no transconjugants obtained after three conjugation assays. Simultaneously, the outcome of oriTfinder analysis revealed that no origin site of DNA transfer (oriT) was predicted (Figure S2).
Existence of Two Accompanying Plasmids p4717_1 and p4717_2
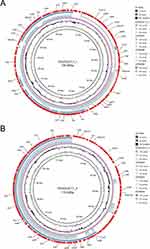
In addition to the p4717-OXA-484, two accompanying plasmids were also found in the clinical strain K. variicola 4717 and designated as p4717_1 and p4717_2. The p4717_1 was classified as IncFII/IncFIBK-type hybrid plasmid which showed high similarity with three known plasmids restricted to K. variicola, namely pKP91 (no. CP000966), p15WZ-82_res (no. CP032357) and pM142-3 (no. CP063868). Surprisingly, no acquired resistance genes were carried by p4717_1 (Figure 3A).
The PlasmidFinder analysis revealed that p4717_2 had no replicons, indicating the plasmid was untypable. The backbone and structure of p4717_2 was similar to that of pBSI138_P3 (no. MT269850), plasmid p4_1_2.1 (no. CP023840) and plasmid pDA33140-112 (no. CP029583) (Figure 3B). In comparison to p4717-OXA-484, p4717_2 was found to contain a greater variety of antibiotic resistance genes, including dfrA1 (trimethoprim), sul1 (sulphonamide), qnrS1 (quinolone), tet(A) (tetracycline), qacE (disinfectant) and blaDHA-1 (β-lactam). Obviously, the existence of ARGs in p4717_2 partly explains the phenotypic resistance of the clinical isolate K. variicola 4717.
Discussion
In comparison to K. pneumoniae, K. variola strains containing carbapenemases resistance genes have not been given as much attention. In our study, the identification of K. variicola carrying blaOXA-484 suggested the wider dissemination than previously thought. Initially, the resistance profile of K. variicola in the environment was extensively researched. In 2017, a K. variicola isolate producing NDM-9 was identified from an urban river in South Korea. Later the presence of beta-lactamase blaTEM-116 in K. variicola was reported from an urban riverine environment in India.21 Other studies also have demonstrated that K. variicola is a pathogenic organism in animals. In Brazil, a polymyxin-resistant K. variicola was collected from birds.22 A previous study described the isolation of K. variicola in a horse with respiratory disease for the first time.23 K. variicola has been identified as a cause of clinical mastitis in cows, with evidence of its presence in milk from affected animals.24 Shen et al suggested that even low infection levels of K. variicola AHKv-S01 can caused a significant reduction in chicken embryo hatchability.25 As well, the circulation of multidrug-resistant K. variicola in healthy livestock animals has also been reported.26
Contrary to the existing studies that investigate the role of K. variicola in environmental resistance and animal disease, there is growing evidence of its clinical impact. Previous research has demonstrated that K. variicola is associated with a range of infections, including bloodstream infections, urinary infections, and respiratory tract infections. Akine et al has reported a case of meningitis caused by K. variicola after neurosurgery, and the effect of K. variicola has been underestimated in terms of causing urinary tract infection disease.3,27 Studies have demonstrated that bloodstream infections caused by K. variicola are more fatal than K. pneumoniae, and this pathogen is mainly isolated from blood samples in clinical settings.28 Moreover, K. variicola and K. pneumoniae isolated from the same patient had the different antimicrobial susceptibility profiles for K. variicola was imipenem-susceptible, but K. pneumoniae was resistant.29,30 However, K. variicola 4717 isolated in this work showed resistance to both imipenem and meropenem. The biofilm formation assay showed K. variicola 4717 was a moderate biofilm producer due to the expression of fimbriae factors, granting it the ability to adhere and colonize. Understanding the relevant pathogenic and resistant characteristics of K. variicola was indispensable, which contributes to the identification and clinical treatment of associated infections.
The blaKPC gene is the most commonly reported gene conferring carbapenem resistance in K. variicola, while the OXA-48-like carbapenemase is relatively uncommon. In a retrospective study, only one strain carrying blaOXA-48 was detected in 145 K. variicola strains.6 It also has been suggested that the blaOXA-484-bearing strains had a lower resistance to temocillin and carbapenems than the blaOXA-48-carrying strains.9 While, there were few reports on carbapenem resistance genes-harboring K. variicola in China. In contrast to previous reports about blaOXA-484, the discovery of K. variicola 4717 complements a novel example of blaOXA-484-positive strain. Utilizing the genome sequence of K. variicola 4717 as a reference point for comparative genomic analysis can provide further insight into the transmission mechanism of K. variicola.
Plasmids are often seen as a crucial genetic factor in the transfer of antibiotic resistance genes between different species.31,32 The p4717-OXA-484 belonged to the IncX3 plasmids which were particularly renowned for their broad host range and high transmissibility.33,34 But the results of the oriTfinder analysis indicated that p4717-OXA-484 may not be capable of conjugation and transfer due to the lack of oriT modules, which was in line with the unsuccessful results of the conjugation transfer experiment. It was likely that the blaOXA-484 gene, which encodes for carbapenemase in K. variicola 4717 is a result of mutation from the blaOXA-181 gene that is present in E. coli and K. pneumoniae isolates, given that the genetic structures are analogous.
Conclusion
Our initial discovery of a K. variicola strain harboring the blaOXA-484 gene encouraged us to explore its resistance mechanism and genetic features. Our research shed light on the current prevalence of ARGs and the importance of bacterial identification. Paying close attention to K. variicola and monitor its carriage of ARGs is critical to prevent it from developing into a novel pathogen.
Ethics Approval
The ethical protocol was approved by the Ethics Committee of First Affiliated Hospital of Zhejiang University (no. 2021-631).
Acknowledgments
This work was supported by research grants from Henan Science and Technology Department (192102310059), the National Natural Science Foundation of China (82072314), the Research Project of Jinan Microecological Biomedicine Shandong Laboratory (JNL-2022011B), the Fundamental Research Funds for the Central Universities (2022ZFJH003), CAMS Innovation Fund for Medical Sciences (2019-I2M-5-045), Henan Province Medical Science and Technology Research Project Joint Construction Project (LHGJ20190232), and Zhejiang Provincial Natural Science Foundation of China (LQ20H200003).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Martin RM, Bachman MA. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8:4. doi:10.3389/fcimb.2018.00004
2. Rosenblueth M, Martínez L, Silva J, Martínez-Romero E. Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst Appl Microbiol. 2004;27(1):27–35. doi:10.1078/0723-2020-00261
3. Rodríguez-Medina N, Barrios-Camacho H, Duran-Bedolla J, Garza-Ramos U. Klebsiella variicola: an emerging pathogen in humans. Emerg Microbes Infect. 2019;8(1):973–988. doi:10.1080/22221751.2019.1634981
4. Nakamura-Silva R, Macedo LMD, Cerdeira L, Oliveira-Silva M, Silva-Sousa YTC, Pitondo-Silva A. First report of hypermucoviscous Klebsiella variicola subsp. variicola causing primary endodontic infection. Clin Microbiol Infect. 2021;27(2):303–304. doi:10.1016/j.cmi.2020.07.045
5. Wang B, Pan F, Han D, et al. Genetic characteristics and microbiological profile of hypermucoviscous multidrug-resistant Klebsiella variicola coproducing IMP-4 and NDM-1 carbapenemases. Microbiol Spectr. 2022;10(1):e0158121. doi:10.1128/spectrum.01581-21
6. Potter RF, Lainhart W, Twentyman J, et al. Population structure, antibiotic resistance, and uropathogenicity of Klebsiella variicola. mBio. 2018;9(6). doi:10.1128/mBio.02481-18
7. Sattler J, Tsvetkov T, Stelzer Y, et al. Emergence of Tn1999.7, a new transposon in bla(OXA-48)-harboring plasmids associated with increased plasmid stability. Antimicrob Agents Chemother. 2022;66(11):e0078722. doi:10.1128/aac.00787-22
8. Pitout JDD, Peirano G, Kock MM, Strydom KA, Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev. 2019;33(1). doi:10.1128/CMR.00102-19
9. Sommer J, Gerbracht KM, Krause FF, et al. OXA-484, an OXA-48-type carbapenem-hydrolyzing class D β-lactamase from Escherichia coli. Front Microbiol. 2021;12:660094. doi:10.3389/fmicb.2021.660094
10. Moser AI, Campos-Madueno EI, Sendi P, et al. Repatriation of a patient with COVID-19 contributed to the importation of an emerging carbapenemase producer. J Glob Antimicrob Resist. 2021;27:267–272. doi:10.1016/j.jgar.2021.10.012
11. Werinder A, Aspán A, Söderlund R, et al. Whole-genome sequencing evaluation of MALDI-TOF MS as a Species Identification Tool for Streptococcus suis. J Clin Microbiol. 2021;59(11):e0129721. doi:10.1128/JCM.01297-21
12. Zhao J, Zheng B, Xu H, et al. Emergence of a NDM-1-producing ST25 Klebsiella pneumoniae strain causing neonatal sepsis in China. Front Microbiol. 2022;13:980191. doi:10.3389/fmicb.2022.980191
13. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing.
14. Chi X, Zhang J, Xu H, et al. Emergence of KPC-2-producing raoultella ornithinolytica isolated from a hospital wastewater treatment plant. Antimicrob Agents Chemother. 2020;64(2). doi:10.1128/AAC.01983-19
15. Zhang S, Cahalan MD, Purifying plasmid DNA from bacterial colonies using the QIAGEN miniprep kit. J Vis Exp. 2007;6:247. doi:10.3791/247
16. Long SW, Linson SE, Ojeda Saavedra M, et al. Whole-genome sequencing of human clinical Klebsiella pneumoniae isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere. 2017;2(4). doi:10.1128/mSphereDirect.00290-17
17. Bortolaia V, Kaas RS, Ruppe E, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–3500. doi:10.1093/jac/dkaa345
18. Barrios-Camacho H, Aguilar-Vera A, Beltran-Rojel M, et al. Molecular epidemiology of Klebsiella variicola obtained from different sources. Sci Rep. 2019;9(1):10610. doi:10.1038/s41598-019-46998-9
19. Llosa M, Gomis-Rüth FX, Coll M, de la Cruz FDF. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol. 2002;45(1):1–8. doi:10.1046/j.1365-2958.2002.03014.x
20. Zou H, Berglund B, Xu H, et al. Genetic characterization and virulence of a carbapenem-resistant Raoultella ornithinolytica isolated from well water carrying a novel megaplasmid containing bla(NDM-1). Environ Pollut. 2020;260:114041. doi:10.1016/j.envpol.2020.114041
21. Mondal AH, Siddiqui MT, Sultan I, Haq QMR. Prevalence and diversity of blaTEM, blaSHV and blaCTX-M variants among multidrug resistant Klebsiella spp. from an urban riverine environment in India. Int J Environ Health Res. 2019;29(2):117–129. doi:10.1080/09603123.2018.1515425
22. Lenzi MH, Martins WM, Roch M, et al. A new mutation in mgrb mediating polymyxin resistance in Klebsiella variicola. Int J Antimicrob Agents. 2021;58(5):106424. doi:10.1016/j.ijantimicag.2021.106424
23. Mondo E, Rinnovati R, Spadari A, et al. First isolation of Klebsiella variicola from a horse pleural effusion. BMC Vet Res. 2021;17(1):75. doi:10.1186/s12917-021-02776-2
24. Podder MP, Rogers L, Daley PK, Keefe GP, Whitney HG, Tahlan K. Klebsiella species associated with bovine mastitis in Newfoundland. PLoS One. 2014;9(9):e106518. doi:10.1371/journal.pone.0106518
25. Shen X, Yin L, Ma H, et al. Comprehensive genomic analysis and characterization of a new ST 174 type Klebsiella variicola strain isolated from chicken embryos. Infect Genet Evol. 2021;90:104768. doi:10.1016/j.meegid.2021.104768
26. Giannattasio-Ferraz S, Ene A, Johnson G, et al. Multidrug-resistant Klebsiella variicola isolated in the urine of healthy bovine heifers, a potential risk as an emerging human pathogen. Appl Environ Microbiol. 2022;88(9):e0004422. doi:10.1128/aem.00044-22
27. Akine D, Sasahara T, Watanabe S, et al. Post-surgical meningitis caused by Klebsiella variicola. IDCases. 2019;18:e00622. doi:10.1016/j.idcr.2019.e00622
28. Imai K, Ishibashi N, Kodana M, et al. Clinical characteristics in blood stream infections caused by Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae: a comparative study, Japan, 2014–2017. BMC Infect Dis. 2019;19(1):946. doi:10.1186/s12879-019-4498-x
29. Garza-Ramos U, Moreno-Dominguez S, Hernández-Castro R, et al. Identification and characterization of imipenem-resistant Klebsiella pneumoniae and susceptible Klebsiella variicola isolates obtained from the same patient. Microb Drug Resist. 2016;22(3):179–184. doi:10.1089/mdr.2015.0181
30. Huang L, Fu L, Hu X, et al. Co-occurrence of Klebsiella variicola and Klebsiella pneumoniae both carrying bla (KPC) from a respiratory intensive care unit patient. Infect Drug Resist. 2021;14:4503–4510. doi:10.2147/IDR.S330977
31. Pu Q, Fan XT, Li H, An XL, Lassen SB, Su JQ. Cadmium enhances conjugative plasmid transfer to a fresh water microbial community. Environ Pollut. 2021;268(Pt B):115903. doi:10.1016/j.envpol.2020.115903
32. Kern-Zdanowicz I. pCTX-M3-structure, function, and evolution of a multi-resistance conjugative plasmid of a broad recipient range. Int J Mol Sci. 2021;22(9). doi:10.3390/ijms22094606
33. Kim JS, Jin YH, Park SH, et al. Horizontal transfer of bla(NDM-1)-carrying IncX3 plasmid between carbapenem-resistant Enterobacteriaceae in a single patient. J Infect. 2020;81(5):816–846. doi:10.1016/j.jinf.2020.07.013
34. Peirano G, Chen L, Nobrega D, et al. Genomic epidemiology of global carbapenemase-producing Escherichia coli, 2015–2017. Emerg Infect Dis. 2022;28(5):924–931. doi:10.3201/eid2805.212535
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.