Back to Journals » Infection and Drug Resistance » Volume 15
Emergence of optrA-Mediated Linezolid Resistance in Enterococcus faecium: A Molecular Investigation in a Tertiary Hospital of Southwest China from 2014–2018
Authors Yi M, Zou J, Zhao J, Tang Y, Yuan Y, Yang B, Huang J, Xia P, Xia Y
Received 14 October 2021
Accepted for publication 14 December 2021
Published 4 January 2022 Volume 2022:15 Pages 13—20
DOI https://doi.org/10.2147/IDR.S339761
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Miao Yi,1 Jiaqi Zou,2 Jinxin Zhao,1 Yu Tang,3 Yaling Yuan,1 Bingxue Yang,1 Jinzhu Huang,1 Peiwen Xia,1 Yun Xia1
1Department of Laboratory Medicine, The First Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China; 2Department of Clinical Laboratory, Chengdu Women’s and Children’s Central Hospital, School of Medicine, University of Electronic Science and Technology of China, Chengdu, People’s Republic of China; 3Department of Laboratory Medicine, Chongqing University Three Gorges Hospital, Chongqing, People’s Republic of China
Correspondence: Yun Xia
Department of Laboratory Medicine, The First Affiliated Hospital of Chongqing Medical University, 1 Youyi Road, Chongqing, 400016, People’s Republic of China
Tel +86-23-89012742
Fax +86-23-89012513
Email [email protected]
Purpose: To investigate the potential mechanism and molecular characteristics of linezolid-non-sensitive Enterococcus faecium from a tertiary hospital in southwest China and characterize the relevant plasmids.
Patients and Methods: Linezolid-non-sensitive Enterococcus faecium (LNSEFM) isolates collected from January 2014 to December 2018 were screened for resistant genes 23s rRNA, rplC, rplD, rplV, optrA, cfr, poxtA, by PCR. Molecular epidemiological analysis was performed by multilocus sequence typing (MLST). The optrA-and-poxtA co-harboring strain EFM_7150 was subjected to the whole genome sequencing (WGS) by Illumina HiSeq and Oxford Nanopore MinION.
Results: A total of 15 LNSEFM with linezolid MICs ranging from 4 to 16 mg/L were identified. About 66.7% (10/15) of isolates were linezolid-resistant. About 46.7% (7/15) of strains were positive for optrA. Two types of optrA variants (P and EYDNDM) were identified. About 13.3% (2/15) of isolates had poxtA. 1 harbored a L22 protein alteration (Ser77Thr). One isolate coharbored optrA (EYDNDM variant) and poxtA. There was no mutation in the gene that encoded the ribosomal protein L3/L4 or the domain V of 23S rRNA. No cfr gene was detected. Based on WGS data, optrA was associated with Tn558 inserted to radC gene and poxtA was flanked by IS1216E.
Conclusion: OptrA is primary mechanism in linezolid-resistant Enterococcus faecium. This is the first report ofoptrA variants P and EYDNDM identified in Enterococcus faecium and optrA-and-poxtA co-harboring Enterococcus faecium clinically in southwest China. Besides, Tn558 and IS1216Es may play an important role in the dissemination of optrA and poxtA, respectively. The findings revealed the potential threat to nosocomial infection by optrA and coexistence of optrA and poxtA in Enterococcus faecium. Thus, clinical surveillance of linezolid-resistant Enterococcus is urgently needed.
Keywords: optrA, poxtA, linezolid-non-sensitive Enterococcus faecium, WGS
Introduction
Enterococcus is a Gram-positive opportunistic pathogen, normally residing in the gastrointestinal tracts of humans, which is regarded as the most common cause of nosocomial infections, such as meningitis, bacteremia, pneumonia, surgical wound infection, and urinary tract infection.1 Its strong intrinsic and acquired resistance leads to resistance to a major group of antibiotics such as vancomycin. Hence, a better understanding of the mechanism of resistance and transmission could support better strategies to monitor and control drug resistance.
The oxazolidinone linezolid targeted at the large (50S) subunit of bacterial ribosomes is considered as the last resort to methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus (VRE), and other multi-drug Gram-positive bacteria.2 However, linezolid-resistant isolates have been increasingly monitored since their clinical use in 2000.3,4 Linezolid-resistant Enterococcus (LRE) represents a significant threat to clinical treatment. The most common resistance mechanism is mutations in the central loop of the domain V region of the 23S rRNA gene, especially G2576T and varying copy numbers.5 Moreover, mutation or deletion of genes that encode the 50S ribosomal subunit proteins L3, L4, L22 and acquisition of resistance genes such as optrA, poxtA also lead to increased linezolid MIC.6–8 Oxazolidinone and phenicol transferable gene optrA was first detected in a clinical Enterococcus faecalis from China in 2015.7 Following its first report, optrA has also been discovered in many countries such as Colombia, Tunisia, Poland.9–11 It is mostly reported in Enterococcus but also detected in Staphylococcus sciuri, Streptococcus suis, and other Gram-positive or Gram-negative strains.12 OptrA is often located on chromosomes or plasmids and can be transmitted by mobile genetic elements such as transposons and insertion sequences.13–15 The most recently reported poxtA gene, mediating resistance to oxazolidinones, tetracyclines, and phenicols, was first described in an MRSA isolate from respiratory secretion of an Italian patient.8 Since its first report, it has also been detected in isolates from animals and humans in many countries, including Italy, Greece, and China.8,16,17 Otherwise, poxtA is found more in the environment or food-producing animals than human samples and Enterococcus faecium has higher prevalence than Enterococcus faecalis.
Our previous transcriptomics and proteomics studies showed that optrA played an important role in linezolid-resistance E. faecalis at the First Affiliated Hospital of Chongqing Medical University and that sexual pheromones could promote optrA transmission.18–21 However, the mechanism of linezolid in E. faecium is not yet clear. Therefore, the purpose of this study is to investigate the mechanism of linezolid in E. faecium and reveal its transmission by whole-genome sequencing. To the best of our knowledge, we monitored the emergence of optrA-mediated linezolid resistance in E. faecium. Besides, this is the first report of optrA variants, P and EYDNDM identified in E. faecium, and optrA and poxtA co-exist in the same strain in southwest China.
Materials and Methods
Bacterial Strains and Antimicrobial Susceptibility Tests
A total of 1891 E. faecium strains were obtained at the First Affiliated Hospital of Chongqing Medical University from January 2014 to December 2018. Excluding duplicate strains, 15 LNSEFMs were collected from six types of samples, including urine, blood, secretion, seroperitoneum, drainage, and bile. Then, they were stocked at −80°C with glycerol. Antimicrobial susceptibility tests were initially confirmed by AST-GP67 cards (BioMérieux) on the VITEK-2 Compact system (bioMérieux, Lyon, France), including linezolid (LZD), clindamycin (CLI), dalfopristin (DAF), tetracycline (TET), erythromycin (ERY), ciprofloxacin (CIP), moxifloxacin (MOX), levofloxacin (LEV), vancomycin (VAN), ampicillin (AMP), penicillin (PEN), tigecycline (TIG), streptomycin (STR), and gentamicin (GEN), and then linezolid MIC is manually reconfirmed by the broth microdilution method. All results were determined according to the CLSI guidelines,22 and the E. faecalis ATCC29212 was used as a reference strain.
DNA Extraction and Molecular Detection of Mutation and Resistance Genes
Following the manufacturer’s protocol, genomic DNA was extracted from bacteria cultured in the logarithmic growth phase using the HiPure Bacterial DNA Kit (Magen, Guangzhou, China). To investigate the mechanism of linezolid resistance, the mutation of 23s rRNA ribosomal proteins L3 (rplC), L4 (RplD), L22 (rplV) and the presence of cfr, optrA, and poxtA were identified using a previously described method.18 Primers and reaction conditions are shown in Supplementary Table S2. All positive PCR products were sent to Sangon Biotech (Shanghai) Co., Ltd. for bidirectional sequencing and blasted against the NCBI nucleotide database. Nucleotides of 23S rRNA and amino acid sequences of L3, L4, and L22 were compared with the reference E. faecium Aus0004 (GenBank Accession No. CP003351) using DNASTAR package MegAlign (Version 7.1.0). The optrA sequence was compared with plasmid pE349 (GenBank Accession No. NG_048023.1).
Multi-Locus Sequence Typing (MLST)
According to the E. faecium MLST scheme, 7 housekeeping genes (adk, pstS, gyd, purK, gdh, ddl, atpA) were amplified using multi-locus sequence typing (MLST) for an analysis of sequences on the Enterococcus faecium database (https://pubmlst.org/mlst). Primers are listed in Supplementary Table S2.
Whole Genome Sequencing (WGS) and Bioinformatic Analysis
Whole-genome sequencing and bioinformatic analysis on optrA-and-poxtA co-harboring isolate EFM-7150 were conducted by Shanghai Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). Following the manufacturer’s protocol, Wizard® Genomic DNA Purification Kit (Promega) was used to extract genomic DNA. Genomic DNA was sequenced using a combination of Illumina and Nanopore sequencing platforms. For Illumina sequencing, 5 μg of genomic DNA was used for library construction. An Illumina HiSeq X Ten with 2 × 150 bp paired-end reads (Illumina) was used to sequence those libraries. For Nanopore sequencing, Covaris G-TUBE (Covaris, MA) was used for spinning 15 μg of genomic DNA to cut the genomic DNA into ∼10 kb fragments, followed by magnetic bead purification and sequencing adapter connection to both ends. After removing low-quality reads, the following reads were assembled into a contig using HGAP and canu.23 In the end, Pilon (1.23) was applied for error correction of Nanopore assembly results. Gene prediction was performed using bioinformatics software Glimmer (3.0) and GeneMarkS.24 Each set of query nucleotide sequences was aligned with NR, Swiss-Prot and Pfam databases. Circular representation of complete plasmids sequences was visualized using the GView server.25 And Plasmid replicons were identified using PlasmidFinder (2.1) (https://cge.cbs.dtu.dk/services/PlasmidFinder/).
Nucleotide Sequence Accession Number
Nucleotide sequences of EFM_7150 complete chromosome and two key plasmids pEF7150-3, pEF7150-5 were collected in GenBank under accession numbers CP079927, CP079928, CP079929, respectively.
Clinical Data
Patient demographics and clinical data including patient age, sex, date, sample, wards, and antibiotic usage were collected from the hospital information system (HIS) and laboratory information system (LIS).
Ethical Approval Statements
This retrospective study was approved by the Evaluation Committee and the Biomedical Ethics Committee of Chongqing Medical University (2021–515). In light of the retrospective and anonymous nature of the study, the Ethics Committee did not require written informed consent provided by participants.
Results
Clinical Information and Antimicrobial Susceptibility Testing
A total of 15 nonduplicated LNSEFM isolates were recovered from 1891 Enterococcus according to VITEK-2 Compact by the BMD method (Supplementary Table S1). Table 1 shows that 15 isolates were obtained from eleven different wards, the most common is gastrointestinal surgery (n = 3) and urology surgery (n = 3), and there is only one isolate in other wards. Urine is the most common source, followed by secretion. According to clinical data, 10 patients were discharged, and 5 patients were transferred. Penicillin, aminoglycosides, carbapenems and cephalosporins were used during the treatment period.
 |
Table 1 Clinical Information and Screening of Linezolid-Resistant Mechanism |
The 15 strains of LNSEFM linezolid MIC spread from 4 to 16 mg/L, of which 5 strains were intermediary, and 10 strains were resistant to linezolid. In addition, the highest drug resistance rate is penicillin (93.3%), erythromycin (93.3%), and ampicillin (93.3%), followed by levofloxacin (86.7%), moxifloxacin (86.7%), and ciprofloxacin (86.7%). All strains are sensitive to vancomycin and tigecycline (Supplementary Table S1).
Screening of Linezolid-Resistant Mechanism and Sequence Type (STs)
The most common mechanism of linezolid resistance in Enterococcus faecium is 23S rRNA gene mutation, but the presence of resistance genes was found to be the main cause in this study.26 The molecular mechanism of linezolid resistance in 15 LNSEFM is detected in Table 1, showing that the positive rate of optrA gene is 46.7%, while the detection rate of poxtA is 13.3%. Of these, one strain carried both poxtA and optrA. The majority of LR E. faecium (70%, 7/10) had optrA. Compared with pE394, two of optrA proteins are wild-type. Four strains had a single point mutation T481P (P variant), and EFM_7150 had K3E, N12Y, Y176D, D247N, G393D, I622M (EYDNDM variant). One LR E. faecium had a L22 protein alteration. No isolates contained cfr gene and L3/L4 alteration. No genetic mechanism was identified in 6 isolates (40%), among which 5 were LI E. faecium. As shown in Table 1, a total of 9 sequence types (ST) were identified among 15 isolates, of which 3 isolates were new ST with 1 to 2 alle mutation. Besides, 46.7% isolates belonged to CC17 clone complex.
Characteristics of Plasmids and the Genetic Environment of optrA and poxtA
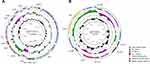
The complete genome of Enterococcus faecium_7150 was constructed using the data from the Illumina HiSeq and Oxford Nanopore MinION. The complete genome has 3,083,859 bp nucleotide and 307,877 reads with a GC content of 37.93%. The plasmids carrying optrA or poxtA are shown in Figure 1. The majority of CDSs code gene in the forward orientation are shown in Supplementary Tables S3 and S4. pEF7150-3 with the GC content of 36.91% has a size of 72,048 bp and 85 Coding sequences were identified. While pEF7150-3 is not completely identical to any other plasmid in the Gene Bank at the moment, as shown from the result of blast analysis, the region around 16,790 bp containing the optrA and fexA genes demonstrated 99.9% similarity and a query coverage of 22% to Enterococcus avium C674 (GenBank accession no. MH018573.1), Staphylococcus sciuri S49-1 optrA gene cluster (GenBank accession no. KX447572.1). Tn558 mediating fexA gene transfer was identified in pEF7150-3 and chromosome. Tn558 and optrA were inserted downstream of the radC gene (encoding a DNA repair protein). Moreover, the transcriptional regulator gene araC was located upstream of optrA.
pEF7150-5 with GC content of 36.11% is 21,754 bp in size and has 22 CDSs. It belongs to rep2 family and the incompatibility (Inc) 18 group plasmids.27,28 pEF7150-5 shared 100% identity with a query coverage of 100% to pM16/0594 (GenBank accession no. MN831411.1), pC25-1 (GenBank accession no. MH784601.1), pC27-2 (GenBank accession no. MH784602.1), pHN11 (GenBank accession no. CP038176.1). Moreover, on plasmid pEF7150-5, the tetracycline resistance gene tet (L) and tet (M) were also identified. The poxtA gene was flanked by two IS1216E elements in the same orientation, which is responsible for horizontal gene transfer of poxtA, as previously described in S. aureus AOUC-0915, E. faecium Efa-955.8,29
Discussion
Linezolid is an effective drug for treating multidrug-resistant Gram-positive bacterial infection. However, the rate of linezolid-resistant has steadily risen in recent years, posing a threat to public health. Zyvox® Annual Appraisal of Potency and Spectrum (ZAAPS), Linezolid Experience and Accurate Determination of Resistance (LEADER), which monitor global pathogens and the changes in resistance to linezolid over time, suggest that the rates of linezolid-non-sensitive were 0.70% and 0.74%, respectively.30,31 In our study, the 5-year prevalence rate of linezolid-resistant E. faecium in our hospital was 1.9%, which is higher than that of China (1.0%).32 Most strains (93.3%) display a multidrug-resistant phenotype (resistant to at least three antimicrobial categories), but all strains are susceptible to vancomycin. It should be noted that nearly half of the isolates belong to CC17 clone that has posed a potential threat to public health, resulting in restricted treatment options worldwide. On antibiotics usage, infected patients had no history of linezolid use during hospitalization, indicating that LNSEFM infection was not associated with linezolid use.
It is well known that mutations in the domain V of the 23S rRNA are the most common mechanism of linezolid resistance among E. faecium, and optrA appears to be almost ubiquitous among the linezolid-non-susceptible E. faecalis.26 Surprisingly, we did not find 23S rRNA mutations but a high prevalence of optrA, which is at odds with the Sentry data.33 This discrepancy may be explained by geographical variation. Almost all resistant isolates, except intermediates, have resistance genes. Nonetheless, we fail to find any previously described mediated genes in linezolid resistance in one isolate (MIC 8mg/L). We speculate that the observed phenotype could be related to cell wall thickness or biofilm formation.34 Although it has been shown that L22 protein mutation could reduce linezolid sensitivity by interfering with binding sites, this mutation is uncommon in Enterococcus.6 Ser77Thr was detected in one LR E. faecium in our study, but the relationship between L22 protein mutation and linezolid MIC remains unclear. Increased copy number of 23S rRNA gene resulted in increased resistance expression in the previous study,35 but there is no evident correlation between optrA variants and oxazolidinone mic in Enterococcus.36 Cai et al revealed that different optrA variants and their genetic context have the potential to regulate linezolid MIC at a variety of levels.37 It is still important to investigate other optrA variants and oxazolidinone resistance level. We identified new types of optrA variants, P and EYDNDM in E. faecium. P variant was distributed in Clostridium difficile and Campylobacter jejuni and EYDNDM variants were found in Enterococcus faecalis, Staphylococcus aureus, Staphylococcus sciuri, Enterococcus avium.12 Given that the relationship between optrA variants and linezolid MIC remains unclear, novel optrA variants in E. faecium is of concern.
To the best of our knowledge, mobile genetic elements contribute significantly to the transmission of resistant genes.38 Tn558 carrying fexA gene is located on both the chromosome and the plasmids in EFM_7150. It integrates optrA through the radC gene in pEF7150-3, resulting in a similar genetic environment to many isolates, such as E. avium C674 isolated from the stool samples of healthy populations in Hangzhou and S. sciuri isolated from Sichuan province and Guangdong province.37,39,40 Notably, Fan et al revealed that optrA can exist in methicillin-resistant coagulase-negative staphylococci including S. sciuri S49-1.39 Clinicians need to be alert to the prevalence of optrA in other superbugs such as VRE.
The recently discovered poxtA, a member of the ABC-F proteins family, shares the homology of 32% with optrA. PoxtA was detected more often in the natural environment than in the clinical setting. The IS1216E-PoxtA-IS1216E segment in our study is similar to S. aureus AOUC-0915, and clinical E. faecium from Italy, Spain, indicating that the genetic background of poxtA is relatively single.8,16,41 In addition to the conjugative plasmid pE035 detected in China, Enterococcus harboring both the optrA and poxtA genes were also found in the environment and human samples from Pakistan, Ireland, Spain, France17,42–45 Unexpectedly, there does not seem to be any obvious synergistic effect when optrA and poxtA coexist. Because the MIC of EFM_7150 is not only similar to optrA or poxtA alone isolates in our study but also to E. faecium C10004 isolated from Spain (8mg/L).44 Furthermore, it is higher than strain isolated from Shanghai (0.5mg/L).46 Despite the emergence of co-existence of optrA and poxtA worldwide, limited genetic context was identified and we hope to obtain more genomic data to explain whether there are other mechanisms or negative regulation.
Last but not the least, these results must be taken cautiously and with some limitations in mind. First, since it included just one center and the relative small number LNSEFM, the results of this study may deviate from other sets of studies. Second, the evidence that only one isolate co-harboring optrA and poxtA is not sufficient. Additional sequencing results that reveal the genetic environment will be more convincing. Nonetheless, our findings indicate the emergence of optrA in Enterococcus faecium. Furthermore, optrA and poxtA can coexist in clinical settings and may be transmitted through MGEs. This is real-world clinical experience and providing a useful information for enterococcal infection.
Conclusion
In summary, this research revealed the emergence of optrA in E. faecium, and the prevalence of optrA gene is higher than that of poxtA in Enterococcus faecium from 2014–2018 in our hospital. MGEs, particularly Transposons and insertion sequences may contribute significantly to the dissemination of optrA and poxtA, respectively. Although linezolid is currently effective in the treatment of enterococcal infections, advanced monitoring of changes in the resistance mechanism of linezolid is needed in the future.
Abbreviations
LNSEFM, linezolid-non-sensitive Enterococcus faecium; LR, linezolid resistant; LI linezolid intermediate; MGEs, mobile genetic elements.
Acknowledgments
We thank the clinical laboratory of the Laboratory Department of the First Affiliated Hospital of Chongqing Medical University for providing experimental space and equipment.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Vu J, Carvalho J. Enterococcus: review of its physiology, pathogenesis, diseases and the challenges it poses for clinical microbiology. Front Biol. 2011;6(5):357. doi:10.1007/s11515-011-1167-x
2. Moellering RC. Linezolid: the first oxazolidinone antimicrobial. Ann Intern Med. 2003;138(2):135–142. doi:10.7326/0003-4819-138-2-200301210-00015
3. Mendes RE, Hogan PA, Streit JM, Jones RN, Flamm RK. Zyvox® Annual Appraisal of Potency and Spectrum (ZAAPS) program: report of linezolid activity over 9 years (2004–12). J Antimicrob Chemother. 2014;69(6):1582–1588. doi:10.1093/jac/dkt541
4. Flamm RK, Mendes RE, Hogan PA, Streit JM, Ross JE, Jones RN. Linezolid surveillance results for the United States (LEADER surveillance program 2014). Antimicrob Agents Chemother. 2016;60(4):2273–2280. doi:10.1128/aac.02803-15
5. Bi R, Qin T, Fan W, Ma P, Gu B. The emerging problem of linezolid-resistant enterococci. J Glob Antimicrob Resist. 2018;13:11–19. doi:10.1016/j.jgar.2017.10.018
6. Miller WR, Munita JM, Arias CA. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther. 2014;12(10):1221–1236. doi:10.1586/14787210.2014.956092
7. Wang Y, Lv Y, Cai J, et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother. 2015;70(8):2182–2190. doi:10.1093/jac/dkv116
8. Huang J, Wang M, Gao Y, Chen L, Wang L. Emergence of plasmid-mediated oxazolidinone resistance gene poxtA from CC17 Enterococcus faecium of pig origin. J Antimicrob Chemother. 2019;74(9):2524–2530. doi:10.1093/jac/dkz250
9. Cavaco LM, Bernal JF, Zankari E, et al. Detection of linezolid resistance due to the optrA gene in Enterococcus faecalis from poultry meat from the American continent (Colombia). J Antimicrob Chemother. 2017;72(3):678–683. doi:10.1093/jac/dkw490
10. Freitas AR, Elghaieb H, León-Sampedro R, et al. Detection of optrA in the African continent (Tunisia) within a mosaic Enterococcus faecalis plasmid from urban wastewaters. J Antimicrob Chemother. 2017;72(12):3245–3251. doi:10.1093/jac/dkx321
11. Gawryszewska I, Zabicka D, Hryniewicz W, Sadowy E. Linezolid-resistant enterococci in Polish hospitals: species, clonality and determinants of linezolid resistance. Eur J Clin Microbiol Infect Dis. 2017;36(7):1279–1286. doi:10.1007/s10096-017-2934-7
12. Schwarz S, Zhang W, Du X-D, et al. Mobile oxazolidinone resistance genes in gram-positive and gram-negative bacteria. Clin Microbiol Rev. 2021;34(3):e00188–20. doi:10.1128/CMR.00188-20
13. Chen W, Mandali S, Hancock SP, et al. Multiple serine transposase dimers assemble the transposon-end synaptic complex during IS607-family transposition. eLife. 2018;7. doi:10.7554/eLife.39611
14. Li D, Li XY, Schwarz S, et al. Tn6674 is a novel enterococcal optra-carrying multiresistance transposon of the Tn554 family. Antimicrob Agents Chemother. 2019;63(9):Sep. doi:10.1128/aac.00809-19
15. Chen L, Han D, Tang Z, Hao J, Xiong W, Zeng Z. Co-existence of the oxazolidinone resistance genes cfr and optrA on two transferable multi-resistance plasmids in one Enterococcus faecalis isolate from swine. Int J Antimicrob Agents. 2020;56(1):105993. doi:10.1016/j.ijantimicag.2020.105993
16. Morroni G, Brenciani A, Antonelli A, et al. Characterization of a multiresistance plasmid carrying the optrA and cfr resistance genes from an enterococcus faecium clinical isolate. Front Microbiol. 2018;9:2189. doi:10.3389/fmicb.2018.02189
17. Hao W, Shan X, Li D, et al. Analysis of a poxtA- and optrA-co-carrying conjugative multiresistance plasmid from Enterococcus faecalis. J Antimicrob Chemother. 2019;74(7):1771–1775. doi:10.1093/jac/dkz109
18. Hua R, Xia Y, Wu W, Yang M, Yan J. Molecular epidemiology and mechanisms of 43 low-level linezolid-resistant enterococcus faecalis strains in Chongqing, China. Ann Lab Med. 2019;39(1):36–42. doi:10.3343/alm.2019.39.1.36
19. Yan J, Xia Y, Yang M, et al. Quantitative proteomics analysis of membrane proteins in enterococcus faecalis with low-level linezolid-resistance. Front Microbiol. 2018;9:1698. doi:10.3389/fmicb.2018.01698
20. Hua R, Xia Y, Wu W, Yan J, Yang M. Whole transcriptome analysis reveals potential novel mechanisms of low-level linezolid resistance in Enterococcus faecalis. Gene. 2018;647:143–149. doi:10.1016/j.gene.2018.01.008
21. Zou J, Tang Z, Yan J, et al. Dissemination of linezolid resistance through sex pheromone plasmid transfer in Enterococcus faecalis. Front Microbiol. 2020;11:1185. doi:10.3389/fmicb.2020.01185
22. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing.
23. Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–736. doi:10.1101/gr.215087.116
24. Mark Borodovsky JM. GENMARK: parallel gene recognition for both DNA strands. Comput Chem. 1993;17(2):123–133. doi:10.1016/0097-8485(93)85004-V
25. Petkau A, Stuart-Edwards M, Stothard P, Van Domselaar G. Interactive microbial genome visualization with GView. Bioinformatics. 2010;26(24):3125–3126. doi:10.1093/bioinformatics/btq588
26. Deshpande LM, Castanheira M, Flamm RK, Mendes RE. Evolving oxazolidinone resistance mechanisms in a worldwide collection of enterococcal clinical isolates: results from the SENTRY Antimicrobial Surveillance Program. J Antimicrob Chemother. 2018;73(9):2314–2322. doi:10.1093/jac/dky188
27. Jensen LB, Garcia-Migura L, Valenzuela AJ, Løhr M, Hasman H, Aarestrup FM. A classification system for plasmids from enterococci and other Gram-positive bacteria. J Microbiol Methods. 2010;80(1):25–43. doi:10.1016/j.mimet.2009.10.012
28. Kohler V, Vaishampayan A, Grohmann E. Broad-host-range Inc18 plasmids: occurrence, spread and transfer mechanisms. Plasmid. 2018;99:11–21. doi:10.1016/j.plasmid.2018.06.001
29. Papagiannitsis CC, Tsilipounidaki K, Malli E, Petinaki E. Detection in Greece of a clinical Enterococcus faecium isolate carrying the novel oxazolidinone resistance gene poxtA. J Antimicrob Chemother. 2019;74(8):2461–2462. doi:10.1093/jac/dkz155
30. Mendes RE, Deshpande L, Streit JM, et al. ZAAPS programme results for 2016: an activity and spectrum analysis of linezolid using clinical isolates from medical centres in 42 countries. J Antimicrob Chemother. 2018;73(7):1880–1887. doi:10.1093/jac/dky099
31. Mendes RE, Deshpande LM, Jones RN. Linezolid update: stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist Updat. 2014;17(1–2):1–12. doi:10.1016/j.drup.2014.04.002
32. Hu F, Guo Y, Yang Y, et al. Resistance reported from China antimicrobial surveillance network (CHINET) in 2018. Eur J Clin Microbiol Infect Dis. 2019;38(12):2275–2281. doi:10.1007/s10096-019-03673-1
33. Lee S-M, Huh HJ, Song DJ, et al. Resistance mechanisms of linezolid-nonsusceptible enterococci in Korea: low rate of 23S rRNA mutations in Enterococcus faecium. J Med Microbiol. 2017;66(12):1730–1735. doi:10.1099/jmm.0.000637
34. Tian Y, Li T, Zhu Y, Wang B, Zou X, Li M. Mechanisms of linezolid resistance in staphylococci and enterococci isolated from two teaching hospitals in Shanghai, China. BMC Microbiol. 2014;14(1):292. doi:10.1186/s12866-014-0292-5
35. Marshall SH, Donskey CJ, Hutton-Thomas R, Salata RA, Rice LB. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother. 2002;46(10):3334–3336. doi:10.1128/aac.46.10.3334-3336.2002
36. Hollenbeck BL, Rice LB. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence. 2012;3(5):421–433. doi:10.4161/viru.21282
37. Cai J, Schwarz S, Chi D, Wang Z, Zhang R, Wang Y. Faecal carriage of optrA-positive enterococci in asymptomatic healthy humans in Hangzhou, China. Clin Microbiol Infect. 2019;25(5):
38. Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31(4). doi:10.1128/cmr.00088-17
39. Fan R, Li D, Wang Y, et al. Presence of the optrA gene in methicillin-resistant staphylococcus sciuri of porcine origin. Antimicrob Agents Chemother. 2016;60(12):7200–7205. doi:10.1128/aac.01591-16
40. Sun C, Zhang P, Ji X, et al. Presence and molecular characteristics of oxazolidinone resistance in staphylococci from household animals in rural China. J Antimicrob Chemother. 2018;73(5):1194–1200. doi:10.1093/jac/dky009
41. Moure Z, Lara N, Marín M, et al. Interregional spread in Spain of linezolid-resistant Enterococcus spp. isolates carrying the optrA and poxtA genes. Int J Antimicrob Agents. 2020;55(6):105977. doi:10.1016/j.ijantimicag.2020.105977
42. Wardenburg KE, Potter RF, D’Souza AW, et al. Phenotypic and genotypic characterization of linezolid-resistant Enterococcus faecium from the USA and Pakistan. J Antimicrob Chemother. 2019;74(12):3445–3452. doi:10.1093/jac/dkz367
43. Egan SA, Shore AC, O’Connell B, Brennan GI, Coleman DC. Linezolid resistance in Enterococcus faecium and Enterococcus faecalis from hospitalized patients in Ireland: high prevalence of the MDR genes optrA and poxtA in isolates with diverse genetic backgrounds. J Antimicrob Chemother. 2020;75(7):1704–1711. doi:10.1093/jac/dkaa075
44. Ruiz-Ripa L, Feßler AT, Hanke D, et al. Detection of poxtA- and optrA-carrying E. faecium isolates in air samples of a Spanish swine farm. J Glob Antimicrob Resist. 2020;22:28–31. doi:10.1016/j.jgar.2019.12.012
45. Dejoies L, Sassi M, Schutz S, et al. Genetic features of the poxtA linezolid resistance gene in human enterococci from France. J Antimicrob Chemother. 2021;76(8):1978–1985. doi:10.1093/jac/dkab116
46. Li P, Yang Y, Ding L, Xu X, Lin D. Molecular investigations of linezolid resistance in Enterococci OptrA variants from a hospital in Shanghai. Infect Drug Resist. 2020;13:2711–2716. doi:10.2147/IDR.S251490
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.