Back to Journals » Infection and Drug Resistance » Volume 15
Emergence of Carbapenem-Resistant ST244, ST292, and ST2446 Pseudomonas aeruginosa Clones in Burn Patients in Yunnan Province
Authors Fang Y, Baloch Z , Zhang W, Hu Y , Zheng R, Song Y, Tai W, Xia X
Received 14 December 2021
Accepted for publication 8 March 2022
Published 16 March 2022 Volume 2022:15 Pages 1103—1114
DOI https://doi.org/10.2147/IDR.S353130
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Yue Fang,1,* Zulqarnain Baloch,1,* Wei Zhang,2 Ying Hu,2 Rui Zheng,3 Yuzhu Song,1 Wenlin Tai,2 Xueshan Xia1
1The Affiliated AnNing First Hospital & Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming, Yunnan, 650500, People’s Republic of China; 2The 2nd Affiliated Hospital of Kunming Medical University, Kunming, Yunnan, 650101, People’s Republic of China; 3The First Hospital of Yunnan Province & The Affiliated Hospital, Kunming University of Science and Technology, Kunming, Yunnan, 650034, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yuzhu Song; Xueshan Xia, Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming, Yunnan, 650500, People’s Republic of China, Tel +86-871-65920756, Fax +86-871-65920570, Email [email protected]; [email protected]
Introduction: The prevalence of carbapenem-resistant Pseudomonas aeruginosa is increasing persistently, particularly in burn ward isolates. Here, we investigate the prevalence of carbapenem-resistant Pseudomonas aeruginosa in a burn ward of a provincial-level hospital at Kunming, Yunnan province, China.
Methods: A total of 118 P. aeruginosa strains were isolated from 57 hospitalized patients, and their MICs were measured. Carbapenem-resistant isolates were selected for multilocus sequence typing (MLST). Carbapenem-resistance mechanisms were identified by examining carbapenemase genes and OprD protein and Carba-NP testing. Representative isolates were further characterized by de novo sequencing for carbapenemase molecular background.
Results: Among 118 P. aeruginosa isolates, 54 (54/118,45.8%) were carbapenem-resistant Pseudomonas aeruginosa, and 3 genotypes were found (ST292, ST244, and ST2446). Non-carbapenemase-producing ST292 was the most prevalent ST, followed by ST2446 and ST244. A novel 13-bp oprD deletion was found in the ST292 clone, which formed the truncated outer membrane protein and may cause carbapenem resistance. ST244 and ST2446 harbored blaIMP-45 and blaIMP-87, respectively. blaIMP-45 is located in a megaplasmid, together with aac(6’)-Ib3, blaOXA-1, catB3, qnrVC6, armA, msr(E), mph(E), aph(3’)-Ia, tetC/tetR, aac(6’)-Ib3, floR, mexC-mexD-oprJ, fosA and lead to extensive drug resistance. ST2446 contains a carbapenem-resistant gene blaIMP-87 on the chromosome and is acquired by a novel gene cassette array (blaIMP-87-ant(2”)-Ia-blaOXA-10-aac(6’)-Ib3) of class 1 integron.
Discussion: For the first time, ST244, ST292 and ST2446 are reported emerging in burn patients, with distinctive carbapenem-resistance mechanisms, respectively. The obtained results highlight the need to surveillance carbapenem-resistant isolates in burn patients.
Keywords: P. aeruginosa, carbapenem-resistance, oprD, blaIMP
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is a common clinical opportunistic pathogen that causes life-threatening nosocomial infections. Burn patients are the main targets of nosocomial infections caused by P. aeruginosa, which is a leading cause of burn patients morbidity and mortality.1 The loss of the skin barrier, cellular damage, and tissue destruction due to burning injury can provide a suitable environment for P. aeruginosa accumulation and production.2 Furthermore, the continuous fluid secretion from burn wounds boosts P. aeruginosa reproduction and stimulates the expression of virulence factors.3
Carbapenems are the drug of choice for treating P. aeruginosa infections. However, the prevalence of carbapenem-resistant P. aeruginosa is increasing annually4 and is becoming a global emerging public health issue. Multiple mechanisms, such as target mutations, outer membrane protein porin loss, enzyme production, and multidrug efflux systems, are responsible for drug resistance in P. aeruginosa.5 IMP enzymes, the class B metalloenzyme, was the first carbapenemase detected in P. aeruginosa.6 blaIMP has been described as an acquirable gene of carbapenemase, and its mobility is related to class 1 integrons located in the plasmid.7 P. aeruginosa plasmids have multiple incompatibility (Inc) or replicon types, such as IncF, N, HI2, P2, and are associated with the blaIMP gene propagation.8,9 Mutations in P. aeruginosa outer membrane porin D (OprD) reduce carbapenems sensitivity and lead to carbapenem resistance.10 However, carbapenem resistance due to OprD mutations cannot be horizontally transferred.
Multilocus sequence typing (MLST) is a portable, unambiguous, DNA-sequence-based technique widely used for molecular typing of bacteria.11 According to estimates, ST274, ST244, ST235, ST277, and ST357 are the widely spread STs of P. aeruginosa in China.12 The prevalence of P. aeruginosa STs among Chinese burn patients are distinct, for example, ST360 and ST316 were predominant in 2011 and 2012, while ST111 emerged in 2013 and became the primary ST in 2014.13 Molecular typing has significant potential in clinical implications, which help identify transmission mechanisms and predict potential STs future outbreaks.
Epidemiological characteristics of P. aeruginosa have indeed been reported from various parts of China. However, the prevalence of P. aeruginosa among burn patients in Yunnan is unclear. Additionally, antimicrobial resistance and genetic diversity of P. aeruginosa among burn patients in Yunnan are also lacking. Therefore, the current study explored the prevalence and antimicrobial resistance of P. aeruginosa among burn patients in Yunnan. Moreover, P. aeruginosa isolates were characterized using de novo sequencing.
Materials and Methods
Isolation and Antimicrobial Susceptibility Testing
From December 2014 to August 2015, burn wound secretion, blood, sputum, and urine samples were collected from patients hospitalized in the burns ward of the Second Affiliated Hospital of Kunming Medical University, located in the center of Kunming, the provincial capital of Yunnan province with approximately 44 million inhabitants. Samples were cultured, and P. aeruginosa strains were isolated. The isolates were identified by the VITEK-2 Compact (bioMérieux, Marcy l’Etoile, France).
The minimum inhibitory concentrations (MICs) of 7 antibiotics named imipenem; meropenem, piperacillin, ceftazidime, aztreonam, levofloxacin, and amikacin were assessed by broth-microdilution susceptibility testing method. P. aeruginosa ATCC25923 served as the control according to the Clinical and Laboratory Standards Institute (CLSI) Guidelines. Carbapenem-resistant isolates that showed resistance to imipenem or meropenem were selected.
Molecular Typing of Isolates
P. aeruginosa isolates were subjected to Multilocus sequence typing (MLST) according to the protocols described in the PubMLST (http://pubmlst.org/paeruginosa) database. Purified PCR products were sent to TsingKe Biological Technology (Kunming, China) and bidirectionally sequenced. Sequences were assembled by the SeqMan module of DNASTAR 7.0 (DNASTAR Inc., Madison, WI, USA). Allele profiles and sequence types (STs) were assigned by the MLST database (http://pubmlst.org/paeruginosa).
Antimicrobial-Resistant Genes Screening and Carbapenemase Phenotypic Detection
DNA was extracted withTIANamp Bacterial DNA Kit (Tiangen Biotech Co., Ltd., Beijing, China). The multiple beta-lactam resistant genes, including carbapenemase genes (blaIMP, blaNDM, blaIMI, blaSIM, blaGIM, blaVIM, blaKPC, blaOXA-23, blaOXA-51), ESBL genes (blaSHV, blaCTX-M, and blaGES), and other beta-lactamase genes (blaPAO, blaCMY, blaOXA-50, blaTEM) were amplified by PCR. Sequencing was performed by TsingKe Biological Technology (Kunming, China). All acquired sequences were compared with the Comprehensive Antibiotic Resistance Database (CARD, https://card.mcmaster.ca). Further, the Carba-NP test was performed to verify the existence of carbapenemases.
Identifying the oprD
The oprD genes were amplified using the previously described primers,14 sequenced, and compared with the oprD gene of PAO1 via MEGA version 5.05. Outer membrane proteins were further isolated and separated by SDS-PAGE.15
De Novo Sequencing and Assembled
Genomic DNA of KB-PA_3 (ST2446), KB-PA_F6 (ST292), and KB-PA-F19 (ST244) were extracted and sent to Beijing Novogene Bioinformatics Technology Co., Ltd for sequencing. Low-quality reads were filtered by the SMRT 2.3.0 and the filtered reads were assembled to generate one contig without gaps.
Genome Annotation and Comparative Analysis
The contigs were annotated using PARIC (https://patricbrc.org/), screened for antimicrobial-resistant genes with resFinder (https://cge.cbs.dtu.dk/services/ResFinder/), analyzed for insertion sequences and integron with ISfinder (https://www-is.biotoul.fr/). INTEGRALL (http://integrall.bio.ua.pt/) was used in integron analysis, and Phaster (http://phaster.ca/) was used in screening phages. Both SnapGene software (Insightful Science, www.snapgene.com) and Mauve tools were used for the comparative analysis of chromosomes.
Nucleotide Sequence Accession Numbers
The oprD sequences of ST292, ST2446, and ST244 isolates were submitted to Genbank with accession numbers OL372281, OL372282, and OL372283, respectively. The whole-genome sequences were submitted to GenBank within BioProject PRJNA774784, and the accession numbers for the chromosome and plasmid of KB-PA_F19 were CP086010, CP086011, CP086012, CP086013, CP086014, and CP086015, respectively. The accession numbers for the chromosome and plasmid of KB-PA_3 were CP086016 and CP086017, respectively.
Ethical Approval
This study was approved by Kunming University of Science and Technology and complied with guideline of the Declaration of Helsinki. Because this study only focused on bacteria, and all the clinical isolates were part of routine hospital laboratory procedures, patients informed consent was not required.
Results
The Prevalence of Carbapenem-Resistant P. aeruginosa Isolates
A total of 118 P. aeruginosa strains were isolated from 57 hospitalized patients. Fifty-four isolates (54/118, 45.8%) were carbapenem-resistant P. aeruginosa, obtained as follows 50 belong to burn wounds, 2 from blood, 1 from urine, and 1 from sputum. MLST was used to analyze the genotypic diversity of P. aeruginosa isolates based on 7 housekeeping genes. A total of 3 distinct MLST types were found (Table 1) with ST allele numbers (acsA-aroE-guaA-mutL-nuoD-ppsA-trpE) were ST244 (17-5-12-3-14-4-7), ST292 (109-10-73-3-4-4-3), ST2446 (111-30-64-26-30-59-55). We found that these 3 sequence types belonged to distinct clonal complexes (A clonal complex was composing STs that shared at least any six of the seven gene alleles). All ST244 and ST2446 isolates were resistant to all tested antimicrobials, except ATM. However, 47/49 ST292 isolates were resistant to beta-lactams and quinolone.
 |
Table 1 The Isolation, MIC Range, Carba-NP Test, and PCR Detections of Resistance Genes Distribution Among the Three STs |
Carbapenem-Resistance Mechanisms
The beta-lactamase genes distribution varied according to the ST type (Table 1). ST244 and ST2446 clones carried the carbapenemase genes blaIMP-45 and blaIMP-87, respectively. However, ST292 was negative for all tested carbapenemase genes. blaSHV-1 and blaCMY-2 genes were present only in ST292 isolates. Moreover, all 54 isolates were positive with blaPAO and blaOXA-50 (blaOXA-396 is a subtype of blaOXA-50).
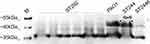
Carba-NP tests were performed to verify isolates may harbor any other carbapenemase. ST292 isolates were negative, while ST244 and ST2446 were positive (Table 1). Further, oprD was found in all carbapenem-resistant P. aeruginosa isolates, and the oprD sequences were identical within the same ST. However, the oprD nucleotide sequences of ST292, ST244, and ST2446 differed from the PAO1 sequence (91% of ST292, 90% of ST2446, and 91% of ST244). Moreover, deletion in oprD at position 233_245del13 of ST292 (Figure 1A) and at the 109del1position of ST2446 were identified. The SDS-PAGE OprD protein profiles of ST292, ST244, and ST2446 showed that ST292 and ST2446 lacked the corresponding band for OprD compared to PAO1 and ST244 (Figure 2).
The General Feature of IMP Producing Genome
Whole-genome sequencing results revealed that KB-PA_3 and KB-PA_F19 genomes consist of single circular chromosomes, with a genome size of 7,098,808-bp and 6,610,918-bp, respectively (Table 2). Compared with the reference chromosome of PAO1 (NC002516.2), KB-PA_3 and KB-PA_F19 contain similar chromosomal information (Figure 3), with different genome architecture. KB-PA_3 chromosome contained ~13% additional nucleotides, and KB-PA_F19 contained ~4.5% additional nucleotide.
 |
Table 2 Chromosome Statistics of P. aeruginosa Strains |
To further analyze the additional sequences, the >4kb additional-regions of KB-PA_3 and KB-PA_F19 were extracted (Supplement S1). KB-PA_F19 had a total of 8 additional regions (max length was ~62k-bp), of which 5 contained phage-related genes and one contained insertion sequence-related genes. KB-PA_3 had 32 additional regions (max length ~109k-bp), of which 15 contained phage-related proteins, 3 contained virulence-related genes (T2SS, fimbria, and hemolysin), 3 contained mobile element genes (insertion sequence and transposon), and one contained T4SS related genes. PHASTER was used to analyze the prophage (Supplement S1), KB-PA_3 contained 7 intact prophage regions and 1 incomplete prophage region. KB-PA_F19 contained 7 intact prophage regions and 2 questionable prophage regions. All predicted prophage regions were located in the additional region of its chromosome. Moreover, KB-PA_3 and KB-PA_F19 shared 2 similar prophage regions (Figure 3), which were predicted as PHAGE_pseudo_F10 (NC007805) and PHAGE_pseudo_PMG1 (NC016765).
KB-PA_3 harbored a ~51k-bp circular plasmid (Table 3). According to analysis, there were a total of 73 CDS in pKB-PA_3-1. Among them, 50 were hypothetical proteins; however, type IV secretion system-related proteins were identified. BLAST analysis showed that pKB-PA_3 was similar to p1011-KPC2 (MH734334.1) with 91% query coverage, p1011-KPC2 was a 62,793-bp plasmid collected from P. aeruginosa in Hangzhou, China. We found that KB-PA_F19 contained five plasmids (range from 1,851-bp to 412,187-bp). Among them, pKB-PA_F19-4 is circular with a genome size of 412,187-bp and considered as megaplasmid (Table 3). The BLAST analysis showed that pKB-PA_F19-4 shares a highly similar backbone with an IncP2-megaplasmid family.16 The other 3 plasmids were linear and may involve gene transmission, mobile elements, and prophages. Even the pKB-PA_F19-2 contained a class 1 integron.
 |
Table 3 Plasmids Identified in P. aeruginosa Strains |
Analysis of Antibiotic Resistance Genes
catB7, fosA, aph(3’)-like, crpP, blaPAO, and blaOXA-50-like genes were located in the chromosome among three P. aeruginosa strains (Table 2), making P. aeruginosa strains naturally resistant to few antimicrobials and with limited resistance level. Sul1, regarded as part of class 1 integron, was also identified in chromosome of KB-PA_3 and KB-PA_F6 isolate. No carbapenemase genes were found in KB-PA_F6; however, aac(6’)-Ib3, aac(6’)-Ib-cr, aadA2b, aph(6)-Id, blaCARB-2, tet(G) genes were identified. The isolates of KB-PA_3 and KB-PA_F19 carried blaIMP genes, which belong to two different families, respectively (Figure 4). blaIMP-87 was located in the chromosome of KB-PA_3 and belonged to the blaIMP-14 family, which involved blaIMP-14, blaIMP-32, and blaIMP-48. The blaIMP-87 protein showed two amino acid substitutions (Val43Ala and Ser47Gly) compared to blaIMP14 and one substitution (Thr69Ile) compared to blaIMP-48. blaIMP-45, belonging to the blaIMP-9 family, which includes blaIMP-9 and blaIMP-53, was located in pKB-PA_F19-4. Moreover, pKB-PA_F19-4 also harbored aac(6’)-Ib3, blaOXA-1, catB3, qnrVC6, armA, Msr(E), Mph(E), aph(3’)-Ia, tetC/tetR, floR, aac(6’)-Ib3, and mexC-mexD-oprJ genes in the same antimicrobial resistance region as blaIMP-45. Fosfomycin resistance protein fosA was identified in another pKB-PA_F19-4 region (156,241 to 158,814) and as associated with ISpa75.
Gene Contexts of blaIMP
blaIMP-87 located within a 5,283-bp class 1 integron (Figure 5), contained a novel gene cassette array (blaIMP-87-ant(2”)-Ia-blaOXA-10-aac(6’)-Ib3). The class 1 integron was flanked by two insertion sequences of IS6100, and then a compound transposon was downstream. The ISPa38-like transposase tnpA was inserted by a complete insertion sequence of IS1071. This carbapenem-resistance region was in the additional region 9 of the chromosome (14,672-bp range from 1,794,528 to 1,795,292, Supplement S1).
The multi-drug resistant megaplasmid pKB-PA_F19-4 shared a similar backbone with its family, and almost all antibiotic-resistance genes were at the plasmid’s resistance region (46,011-bp range from 53,954 to 99,965). BLASTN showed a structural similarity between the pKB-PA_F19-4 resistance region and pBM413 (CP016215, Figure 6). The ARGs and associated mobile elements surrounded the repM and DNA-binding protein gene. Upstream of repM and DNA-binding protein gene, blaIMP-45 is found. blaIMP-45 is composed of a class 1 integron gene cassette array of aac(6’)-Ib3-blaIMP-45-blaOXA-1-cbtB3, and associated with the Tn3-like transposon. Following the class 1 integron, the quinolones resistance gene qnrVC6 is flanked by two ISCR1 elements and then the array of ISEc28-IS1394-armA-ISEc29-msr(E)-mph(E) was identified. Downstream of repM and DNA-binding protein gene, two arrays of IS15-aph (3’)-Ia-IS15 and intI1 acc(6’)-Ib3-ISCfr1-acrR-mexC-mexD-oprJ were identified, showing similar architecture with pBM413. The resistance genes, tetC/tetR and floR, also existed between these two regions.
Discussion
P. aeruginosa is an opportunistic pathogen, commonly infecting hospitalized patients, particularly those in burn wards and ICUs. Moreover, P. aeruginosa has been ranked as one of the most common Gram-negative detected pathogens isolated from burn patients in China.4 In this study, 54 carbapenem-resistant Pseudomonas aeruginosa strains were isolated from patients in the burn ward. P. aeruginosa poses complex acquired and intrinsic carbapenem resistance mechanisms,17,18 which significantly limits the clinician’s choices for antimicrobial selection and treatments.19 Therefore, burn patients infected by carbapenem-resistant P. aeruginosa are at higher risk of infection complications. And phenotypic and molecular characterization of P. aeruginosa isolates from burn patients could provide meaningful information for epidemiology and resistance mechanism.
In this study, ST292 was the most predominant sequence type, followed by ST2446 and ST244. Previously, ST292 has only been reported in clinical isolates of China,12 Thailand,20 and Croatia.21 However, ST292 has never been isolated from burn patients. Here, the high prevalence (41.5%) of ST292 in burn patients is reported for the first time. A long-term outbreak of ST292 was reported in 2016 in a hospital in Southwest China, but the prevalence was not clear.22
The P. aeruginosa porin OprD is a substrate-specific porin that facilitates the diffusion of basic amino acids, small peptides, and carbapenems into the cell (5). Mutation or deletion in OprD has been shown to reduce the susceptibility to carbapenem.21 Here, a novel deletion in OprD at 233_245del13 is reported. This novel deletion may cause early termination before the crystal structures of loop 2 and 3, which contain the entrance and/or binding site for carbapenems and decrease carbapenem susceptibility. We also identified 15 mutations in OprD of the ST292 clone, which may also decrease its susceptibility to carbapenem (Figure 1B).13,21 These results suggest that the ST292 clone obtained carbapenem resistance by mutations or deletions in OprD. Moreover, alteration in OprD can be vertically transmitted within ST292 clones and may lead to enhanced fitness and virulence.10 Our study results suggest that OprD alteration may be allowing ST292 clone to predominate in burn ward patients.
To our best knowledge, we are the first group reporting the ST2446 prevalence in burn and clinical isolates. KB_PAE-3 contains a ~7M-bp chromosome, larger than the average clinical-source P. aeruginosa isolates (~6.6M-bp) and similar to industrial-source isolates (~7M-bp).23 Unusually, T4SS was located in both chromosome and plasmid and function in translocating DNA and protein substrates,24 indicating that KB_PAE-3 genes exchange is more active, leading to many additional sequences in the chromosome.
Additional virulence genes were also detected in KB_PAE-3, increasing the risk of burn patient’s treatment complications. However, it also suggests that the ST2446 clone needs to take advantage of virulence to fight for a fit niche in its original environment. A previous report showed that niche adaptation is a major evolutionary force influencing the composition of P. aeruginosa genomes.25 These results showed that the ST2446 clone was coming from a complex source, most likely animals. Previously, ST2446 has only been reported in Chinese minks causing hemorrhagic pneumonia, but the antimicrobials resistance of ST 2446 remains unknown.26 Furthermore, our study may be the first to report blaIMP-87 in ST2446. Interestingly, IMP-14 and variants were reported as locally spread carbapenemases in Thailand20,27 and once transmitted to Northern Europe.28 During the analysis, we compared this new cassette array with the Thai cassette arrays carried by the blaIMP-14 family (Tn687 and S27-KUU, Figure 6) and found that the blaIMP gene was finally acquired as an independent passenger gene by an integron. A highly similar gene cassette array with 99% identity was present in S27-KUU (KY574887.1) from Thailand, containing blaIMP-48 instead of blaIMP-87. These results suggested that the blaIMP-87-ant(2”)-Ia-blaOXA-10-aac(6’)-Ib3 novel integron cassette harbored by KB_PAE-3 may come from Thailand. Furthermore, blaIMP-14 has a wide host range, except P. aeruginosa.29,30 The blaIMP-87 transposition of the KB_PAE-3 was conducted by IS6100, which has also been reported in E. coli.30 Therefore, it showed that IS6100 is involved in the blaIMP-14 family transmission and may become the driving force for blaIMP cross-species spreading. Together with the oprD 1-bp deletion, the ST2446 clone showed a high carbapenem-resistance with an imipenem MIC over 128 μg/mL. In addition, the role of the plasmid harbored by KB-PA_3 is unknown; however, a similar plasmid carrying blaKPC-2 was reported.31
ST244 has been widely reported globally and listed as a high-risk clone,32 but with few genome characterization. Here, whole-genome sequencing of ST244 isolates was performed. Among carbapenem-resistance genes, blaVIM-2 and blaIMP-6 are prevalent resistance genes in ST244 in China.33,34 However, this study shows that blaIMP-45 is emerging in ST244 and locates in a megaplasmid. Interestingly, when searched in GenBank, the gene cassette arrays of aac(6’)-Ib3-blaIMP-45-blaOXA-1-cbtB3 appeared only in the megaplasmid family. This megaplasmid family has a narrow host range for Pseudomonas spp. Nevertheless, it has been discovered in multiple P. aeruginosa STs, like ST508, ST1420, ST274, and ST708.35 Our results suggest that this type of megaplasmid can transfer among different P. aeruginosa sequence types. Furthermore, this megaplasmid also carried multiple antibiotic resistance genes (Figure 6), such as beta-lactams, carbapenems fluoroquinolone, aminoglycoside, phenicol, sulfonamide, macrolide, lincosamide, streptogramin B, and tetracycline. Different mobile genetic elements carried drug-resistant genes forming this drug-resistant region surrounding the replication site of the megaplasmid, which may also have a role in drug-resistant genes transmission. Furthermore, theISCR1-qnrVC6-ISCR1 array has been reported previously in two Shewanella xiamenensis strains (CP013115.1 and AP025014.1). pBM413 has IS1394 region inserted into the ISCE1-qnrVC6-ISCR1 array. The array of ISEc28-armA-ISEc29-msr(E)-mph(E) was reported in Klebsiella pneumoniae,36 which unlike our study, omits IS1394. The similar resistance architecture between pKB-PA_F19-4 and pBM413 implies that these plasmids may have the same origin. The class 1 integron in pBM413 was truncated class 1 integron In0 without any passenger genes, while pKB-PA_F19-4 carried acc(6’)-Ib3. To the best of our knowledge, tetC/tetR and floR were seldom reported in clinical P. aeruginosa, and these genes may lead to tetracycline and florfenicol resistance, respectively. So far, the megaplasmid family role in antibiotic resistance is unclear, therefore, it is urgent to control the prevalence and transmission of these types of plasmids to avoid the spread of these types of resistance.
Conclusion
Here, the prevalence of carbapenem-resistant P. aeruginosa in the burns ward of the Second Affiliated Hospital of Kunming Medical University was surveyed, showing a unique molecular carbapenem-resistance mechanism in ST292, ST244, and ST2446 clones. Defective OprD mutation formed the ST292 clone most predominant. However, ST244 harbored a multi-drug resistant megaplasmid. ST2446, which has been reported in animals but not in humans, showed high-level carbapenem-resistance. It is important to explore and understand the transmission route of ST244, ST292, and ST2446 in burn patients and design and implement policies to control their spread.
Data Sharing Statement
The data presented in this study are available on request from the corresponding author.
Funding
This work was supported by the Major Scientific Project (2019ZF004) of Yunnan Province.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Nathwani D, Raman G, Sulham K, Gavaghan M, Menon V. Clinical and economic consequences of hospital-acquired resistant and multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2014;3. doi:10.1186/2047-2994-3-32.
2. Coban YK. Infection control in severely burned patients. World J Crit Care Med. 2012;1(4):94. doi:10.5492/wjccm.v1.i4.94
3. Gonzalez MR, Fleuchot B, Lauciello L, et al. Effect of human burn wound exudate on Pseudomonas aeruginosa virulence. mSphere. 2016;1. doi:10.1128/msphere.00111-15
4. Dou Y, Huan J, Guo F, Zhou Z, Shi Y. Pseudomonas aeruginosa prevalence, antibiotic resistance and antimicrobial use in Chinese burn wards from 2007 to 2014. J Int Med Res. 2017;45:1124–1137. doi:10.1177/0300060517703573
5. Hong DJ, Bae IK, Jang IH, Jeong SH, Kang HK, Lee K. Epidemiology and characteristics of metallo-ß-lactamase-producing Pseudomonas aeruginosa. Infect Chemother. 2015;47:81–97. doi:10.3947/ic.2015.47.2.81
6. Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:147–151. doi:10.1128/AAC.35.1.147
7. Zhao WH, Hu ZQ. IMP-type metallo-β-lactamases in gram-negative bacilli: distribution, phylogeny, and association with integrons. Crit Rev Microbiol. 2011;37:214–226. doi:10.3109/1040841X.2011.559944
8. Xiong J, Hynes MF, Ye H, et al. BlaIMP-9 and its association with large plasmids carried by Pseudomonas aeruginosa isolates from the People’s Republic of China. Antimicrob Agents Chemother. 2006;50:355–358. doi:10.1128/AAC.50.1.355-358.2006
9. Liu W, Dong H, Yan T, et al. Molecular characterization of blaIMP–4-carrying enterobacterales in Henan Province of China. Front Microbiol. 2021;12. doi:10.3389/FMICB.2021.626160
10. Skurnik D, Roux D, Cattoir V, et al. Enhanced in vivo fitness of carbapenem-resistant oprD mutants of Pseudomonas aeruginosa revealed through high-throughput sequencing. Proc Natl Acad Sci. 2013;110:20747–20752. doi:10.1073/pnas.1221552110
11. Kidd TJ, Grimwood K, Ramsay KA, Rainey PB, Bell SC. Comparison of three molecular techniques for typing Pseudomonas aeruginosa isolates in sputum samples from patients with cystic fibrosis. J Clin Microbiol. 2011;49(1):263–268. doi:10.1128/JCM.01421-10
12. Ji J, Wang J, Zhou Z, Wang H, Jiang Y, Yu Y. Multilocus sequence typing reveals genetic diversity of carbapenem- or ceftazidime-nonsusceptible Pseudomonas aeruginosa in China. Antimicrob Agents Chemother. 2013;57(11):5697–5700. doi:10.1128/AAC.00970-13
13. Yin S, Chen P, You B, et al. Molecular typing and carbapenem resistance mechanisms of Pseudomonas aeruginosa isolated from a Chinese burn center from 2011 to 2016. Front Microbiol. 2018;9. doi:10.3389/fmicb.2018.01135
14. Rodríguez-Martínez JM, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53(11):4783–4788. doi:10.1128/AAC.00574-09
15. Carlone GM, Thomas ML, Rumschlag HS, Sottnek FO. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J Clin Microbiol. 1986;24:3. doi:10.1128/jcm.24.3.330-332.1986
16. Cazares A, Moore MP, Hall JPJ, et al. A megaplasmid family driving dissemination of multidrug resistance in Pseudomonas. Nat Commun. 2020;11(1):1. doi:10.1038/s41467-020-15081-7
17. Gajdács M, Baráth Z, Kárpáti K, et al. No correlation between biofilm formation, virulence factors, and antibiotic resistance in pseudomonas aeruginosa: results from a laboratory-based in vitro study. Antibiotics. 2021;10(9):9. doi:10.3390/antibiotics10091134
18. Ratajczak M, Kamińska D, Nowak-Malczewska DM, Schneider A, Dlugaszewska J. Relationship between antibiotic resistance, biofilm formation, genes coding virulence factors and source of origin of Pseudomonas aeruginosa clinical strains. Ann Agric Environ Med. 2021;28(2):2. doi:10.26444/aaem/122682
19. Gajdács M, Kárpáti K, Stájer A, Zanetti S, Donadu MG. Insights on carbapenem-resistant Pseudomonas aeruginosa: phenotypic characterization of relevant isolates. Acta Biol Szeged. 2021;65(1):1. doi:10.14232/ABS.2021.1.105-112
20. Khuntayaporn P, Yamprayoonswat W, Yasawong M, Chomnawang MT. Dissemination of carbapenem-resistance among multidrug resistant Pseudomonas aeruginosa carrying metallo-beta-lactamase genes, including the novel bla IMP-65 gene in Thailand. Infect Chemother. 2019;51(2):107. doi:10.3947/ic.2019.51.2.107
21. Guzvinec M, Izdebski R, Butic I, et al. Sequence types 235, 111, and 132 Predominate among multidrug-resistant Pseudomonas aeruginosa clinical isolates in Croatia. Antimicrob Agents Chemother. 2014;58(10):6277–6283. doi:10.1128/AAC.03116-14
22. Fan X, Wu Y, Xiao M, et al. Diverse genetic background of multidrug-resistant Pseudomonas aeruginosa from Mainland China, and emergence of an extensively drug-resistant ST292 clone in Kunming. Sci Rep. 2016;6(1):1–8. doi:10.1038/srep26522
23. Weiser R, Green AE, Bull MJ, et al. Not all Pseudomonas aeruginosa are equal: strains from industrial sources possess uniquely large multireplicon genomes. Microb Genom. 2019;5. doi:10.1099/mgen.0.000276
24. Christie PJ, Lovett ST, Bernstein HD. The mosaic type IV secretion systems. EcoSal Plus. 2016;7:1. doi:10.1128/ecosalplus.esp-0020-2015
25. Mathee K, Narasimhan G, Valdes C, et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A. 2008;105:8. doi:10.1073/pnas.0711982105
26. Bai X, Liu S, Zhao J, et al. Epidemiology and molecular characterization of the antimicrobial resistance of pseudomonas aeruginosa in Chinese mink infected by hemorrhagic pneumonia. Can J Vet Res. 2019;83:2.
27. Prombhul S, Tribuddharat C, Laikijrung P, Aranya C, Bamrungsri N, Mekviwattanawong S. New variant of an imipenemase, IMP-32, in Klebsiella pneumoniae from a fatal case of a Thai patient. J Med Microbiol. 2016;65:6. doi:10.1099/jmm.0.000252
28. Samuelsen Ø, Toleman MA, Sundsfjord A, et al. Molecular epidemiology of metallo-β-lactamase-producing Pseudomonas aeruginosa isolates from Norway and Sweden shows import of international clones and local clonal expansion. Antimicrob Agents Chemother. 2010;54:1. doi:10.1128/AAC.00824-09
29. Kazmierczak KM, Rabine S, Hackel M, et al. Multiyear, multinational survey of the incidence and global distribution of metallo-β-lactamase-producing enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2016;60:2. doi:10.1128/AAC.02379-15
30. Stoesser N, Sheppard AE, Peirano G, et al. First report of blaIMP-14 on a plasmid harboring multiple drug resistance genes in Escherichia coli sequence type 131. Antimicrob Agents Chemother. 2016;60:8. doi:10.1128/AAC.00840-16
31. Hu YY, Wang Q, Sun QL, Chen GX, Zhang R. A novel plasmid carrying carbapenem-resistant gene blakpc-2 in pseudomonas aeruginosa. Infect Drug Resist. 2019;12:1285–1288. doi:10.2147/IDR.S196390
32. Del Barrio-Tofiño E, López-Causapé C, Oliver A. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int J Antimicrob Agents. 2020;56:6. doi:10.1016/j.ijantimicag.2020.106196
33. Chen Y, Sun M, Wang M, Lu Y, Yan Z. Dissemination of IMP-6-producing Pseudomonas aeruginosa ST244 in multiple cities in China. Eur J Clin Microbiol Infect Dis. 2014;33:1181–1187. doi:10.1007/s10096-014-2063-5
34. Feng W, Sun F, Wang Q, et al. Epidemiology and resistance characteristics of Pseudomonas aeruginosa isolates from the respiratory department of a hospital in China. J Glob Antimicrob Resist. 2017;8:142–147. doi:10.1016/j.jgar.2016.11.012
35. Zhang X, Wang L, Li D, et al. An IncP-2 plasmid sublineage associated with dissemination of bla IMP-45 among carbapenem-resistant Pseudomonas aeruginosa. Emerg Microbes Infect. 2021;10:1. doi:10.1080/22221751.2021.1894903
36. Hayashi W, Iimura M, Soga E, et al. Presence of colistin- and tigecycline-resistant Klebsiella pneumoniae ST29 in municipal wastewater influents in Japan. Microb Drug Resist. 2021;27:10. doi:10.1089/mdr.2020.0514
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.