Back to Journals » Infection and Drug Resistance » Volume 13
Emergence of blaNDM-1 Harboring Klebsiella pneumoniae ST29 and ST11 in Veterinary Settings and Waste of Pakistan
Authors Chaudhry TH, Aslam B, Arshad MI , Alvi RF , Muzammil S, Yasmeen N, Aslam MA, Khurshid M , Rasool MH, Baloch Z
Received 2 February 2020
Accepted for publication 17 July 2020
Published 26 August 2020 Volume 2020:13 Pages 3033—3043
DOI https://doi.org/10.2147/IDR.S248091
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Tamoor Hamid Chaudhry,1,2 Bilal Aslam,1,2 Muhammad Imran Arshad,3 Roman Farooq Alvi,2 Saima Muzammil,2 Nafeesa Yasmeen,4 Muhammad Aamir Aslam,3 Mohsin Khurshid,2 Muhammad Hidayat Rasool,2 Zulqarnain Baloch1
1Biomedical Research Center, Northwest Minzu University, Lanzhou 730030, People’s Republic of China; 2Department of Microbiology, Government College University Faisalabad, Faisalabad, Pakistan; 3Institute of Microbiology, University of Agriculture Faisalabad, Faisalabad, Pakistan; 4College of Veterinary Medicine, South China Agricultural University, Guangzhou 510642, People’s Republic of China
Correspondence: Bilal Aslam; Zulqarnain Baloch Email [email protected]; [email protected]
Introduction: Intense livestock farming practices enforcing the farmers to use antibiotics as food supplements on a routine basis. Aberrant use of antibiotics is associated with the emergence of antibiotics resistance and resistant superbugs. Keeping in view the current scenario, the present study was designed for the first time from Pakistan with a specific aim to estimate the prevalence of the carbapenem-resistant Klebsiella pneumoniae in veterinary settings and the waste in Pakistan.
Methods: A total of 138 samples from various veterinary sources were collected by employing a nonprobability sampling technique. Isolation and phenotypic identification of carbapenem-resistant K. pneumoniae were performed according to the CLSI standard. Molecular detection of various antibiotic resistance genes (ARGs) was done through PCR by using specific primers against each ARG. According to the pasture scheme, the multilocus sequence typing (MLST) was performed to characterize the K. pneumoniae sequence types (STs).
Results: According to the results of the study, overall 9.4% (13/138) isolates were confirmed carbapenem-resistant K. pneumoniae. Among various carbapenem ARGs particularly, the blaNDM-1 was found in 92.3% (12/13) isolates followed by blaOXA-48 84.6% (11/13). MLST results revealed that overall 3 STs were found in the study which includes ST29, ST11, and ST258. Taking together, this is the first study to our best knowledge which demonstrated the prevalence of carbapenem-resistant K. pneumoniae and its various STs prevalent in veterinary settings and the waste of Pakistan.
Conclusion: Based on the above-mentioned facts, we suggested that veterinary settings and waste are the potential source and reservoir of carbapenem-resistant K. pneumoniae, which may be disseminated to the environment and ultimately can affect the public and companion livestock health.
Keywords: veterinary settings, carbapenem resistance, Klebsiella pneumoniae, Pakistan
Introduction
The modern intensive integrated livestock production systems require regular antibiotics use at farms to maintain animal health and production. Globally, it is suggested that antibiotic consumption is double in animals compared to humans. The regular and imprudent use of antibiotics in modern veterinary practices is associated with the emergence of different multidrug-resistant (MDR) bacteria.1,2 These MDR pathogens of animal origin may be disseminated to humans via the wider environment including food products, sewage and agricultural system.1–3
Klebsiella pneumoniae is the most important member of the Klebsiella genus of Enterobacteriaceae. It can transfer to humans through contaminated products of like poultry products, beef, fish, milk, etc.4,5 Additionally, the prevalence of carbapenem-resistant K. pneumoniae causing community and hospital-acquired infections is increasing significantly throughout the world, especially in developing countries.6,7 Carbapenems are a class of beta-lactam antibiotics, are regularly prescribed as a last choice in the treatment of Gram-negative bacteria infection but the emergence of carbapenem-resistant Enterobacteriaceae is a critical clinical problem in the world.8,9
The key mechanisms for carbapenem resistance include alteration in porins function or expression, efflux pumps, and acquirement of enzymes capable of antibiotic hydrolyzing called carbapenemases. These enzymes are b-lactamases of Ambler class A, class D, and class B (Metallo-β-lactamases; MBLs).1,10,12–14 Currently, a total of 5 Carbapenemases are key public health concerns, among them, New Delhi Metallo- β-lactamase-1 (NDM-1) has attracted the most attention because blaNDM-1 has diverse antibiotic resistance activity.15 The blaNDM-1 was the first time reported in K. pneumoniae isolated from an Indian patient in Sweden in 2008.8 Since then a lot of studies from various regions of the world have reported the occurrence of the blaNDM-1.8,9 Despite all these reports so far, data about the detection of blaNDM-1 in veterinary and animal settings is still lacking.
Pakistan is a leading livestock populated country in the world. In the last decade, intensive livestock farming increased on a large scale in Pakistan. Therefore, livestock farmers are adapting the modern intense farming protocols in which antibiotics are used as food supplements routinely. This unrestrained antibiotic application in livestock feed is a leading factor that steers the evolution of multidrug-resistant (MDR) pathogens. On the other hand, monitoring practices of antibiotics usage in veterinary and farm settings is inadequate.16 Recently we have reported the first blaKPC harboring K. pneumoniae ST258 from Pakistan and we also have reported the occurrence of ESBL-producing K. pneumoniae in hospital settings and waste from Pakistan.3,17 Here, for the first time from Pakistan, we demonstrated the prevalence of blaNDM-1 harboring carbapenem-resistant K. pneumoniae in veterinary settings and waste from Pakistan.
Materials and Methods
Ethical Approval and Study Settings and Sample Collection
The study was approved by the Ethical Review Board (ERB), Government College University, Faisalabad Pakistan (letter No. 4162 dated 23–11-2017). Sample collection and processing was done at the Department of Microbiology Government College University, Faisalabad Pakistan while, molecular investigations were conducted at the Paul G. Allen School for Global Animal Health, Washington State University, Washington, USA, and Biomedical Research Center, Northwest Minzu University, Lanzhou China. In this study, we applied a nonprobability sampling technique to collect the samples (n= 138). The detail about samples distribution is given in Table 1. Standard aseptic conditions and microbiological procedures were adopted for sample collection and transportation.
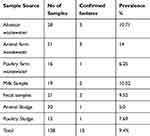 |
Table 1 Description the Sample Distribution and Their Positive Results |
Isolation and Identification of Klebsiella pneumoniae
In brief, abattoir samples and fecal swabs were dipped into 1mL of PBS and then streaked on the surface of nutrient media; however, milk and wastewater samples were directly streaked on nutrient agar plates and were incubated at 37°C for 24 hours. Subsequently, Mackonkey’s agar and HiChrom Klebsiella Selective agar (Himedia®) were used as selective media for the isolation of K. pneumoniae. API 20E Kit and VITEK identification system (bioMérieux, France) was applied for the biochemical identification of the isolates.
Molecular identification was done by amplifying and sequencing the 16S rDNA using specific primers through PCR. The reaction started with the initial melting temperature at 95°C for 3 min, a total of 35 PCR cycles were carried out with following scheme: denaturation at 95°C for 30 secs, annealing at 50°C for 25 secs, extension at 72°C for 65 sec and final extension was done at 72°C for 5 min. Afterward, agarose (ThermoFisher Scientific, USA) gel electrophoresis was performed and results were interpreted in the gel documentation system (Bio-Rad, USA).
Antibiotic Susceptibility Testing
The disc-diffusion assay was used to decipher the antibiotic resistance pattern of the isolates As described in CLSI 2018.18 Various groups of antibiotics along with their concentration used in the antibiogram analysis of K. pneumoniae include Penicillins (ampicillin 10µg and piperacillin 100µg), Cephalosporins (cefuroxime 30µg, cefixime 5µg, ceftriaxone 30µg, and cefepime 30µg), Carbapenems (meropenem 10µg), Fluoroquinolones (ciprofloxacin 5µg), Tetracycline (tetracycline 30µg and minocycline 30µg), Sulfa drugs (trimethoprim-sulfamethoxazole 1.25/23.75µg), Polymyxins (colistin 10µg), and tigecycline 15µg. All the tests were performed in duplicate and a control strain of K. pneumoniae (ATCC® 13,883™) was used for the assay.
For further confirmation, the minimum inhibitory concentration (MIC) of the above-listed antibiotics against the 13 CRKP isolates was determined using broth micro-dilution assay as described previously.12,13 Briefly, fresh isolates of K. pneumoniae were used to obtain 0.5 McFarland standards. Dilutions of the antibiotics were made with the concentrations range 0.06 μg/mL to 256 μg/mL. Inoculum from the wells showing the turbidity was streaked on nutrient agar plates for the confirmation of bacterial growth. The MIC of the isolates showing growth in the well having the antibiotic concentration of 256 μg/mL were considered as ≥256 μg/mL Results were interpreted according to the CLSI recommendations.
Phenotypic Detection of Carbapenem-Resistant K. pneumoniae
The Double Disc Synergy Test (DDST) was employed for the phenotypic characterization of ESBL producing K. pneumonia as described previously.18 Briefly, a 30 μg containing cefotaxime disc alone and a cefotaxime disc in combination with clavulanic acid, having the concentration of 30:10 μg, respectively were placed at a distance of 20 mm, and the difference between discs in terms of zone of inhibition was observed, the difference of ≥5 mm was considered positive for ESBL production.
Modified Hodge Test (MHT) was employed for phenotypic confirmation of carbapenem resistance. A 0.5 McFarland standard dilution of ATCC 25,922 (E. coli) was prepared in broth; a lawn was made using ten-fold dilution. In the center, ertapenem (10 µg) disk was placed. From the edge of the disk, the isolate was streaked up to the edge of the plate. Plates were incubated at 37°C for 24 hours. According to CLSI guidelines, MHT Positive ATCC1705 (K. pneumoniae) and MHT Negative ATCC1706 (K. pneumoniae) were used as control. Besides, MHT positive isolates were confirmed through the CarbNP test as recommended by CLSI 2018.
Molecular Detection of Antibiotic Resistance Genes (ARGs)
Freshly grown isolates of K. pneumoniae were subjected to DNA extraction through the DNA Extraction kit (Qiagen, Hilden, Germany). Quantification of extracted genomic DNA was carried out by using Thermo Scientific™ Nanodrop 2000, USA and DNA ≥60ng/μL concentration were considered further for the experiment. The extracted DNA of all isolates were subjected to PCR for the amplification of ARGs listed in Table 2 by using specific primers (IDT™, USA). The composition of the reaction mixture was; 5 µL of template DNA, 2 µL 100 pM forward and reverse primers, 10 µL of PCR Master Mix (2X DreamTaq, Thermo-Scientific™). The final volume was adjusted to 25 µL with sterile dH2O/Nuclease free water (Ambion-AM9932). The annealing temperature of each gene was adjusted according to primer’s Tm described in Table 2. Subsequently, PCR products were analyzed on 1.2% agarose gel electrophoresis and examined under the gel documentation system (BioRad, USA).
 |
Table 2 Table Showing the Primer Sequence, Amplification Conditions and Distribution of Different Antibiotic-Resistance Determinants Among K. pneumoniae Isolates |
Multi Locus Sequence Typing
The MLST was done according to the Pasteur MLST scheme (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html) by amplification of seven housekeeping genes of K. pneumoniae isolates. The primers for gapA, infB, mdh, pgi, phoE, rpoB and tonB were manufactured from Integrated DNA Technologies Inc. (California, USA). MLST PCR cycle conditions were as follow: denaturation at 94°C for 2 minutes, followed by 35 cycles of initial denaturation at 94°C for 20 seconds, annealing for all genes at 50° expect for gapA (60°) and tonB (45°) for 30 seconds and extension at 72°C for 30 seconds and a final extension at 72°C for 5 minutes in PCR Thermal Cycler (Bio-Rad Inc., USA).19
Conjugation Assay
Plasmid transferability was determined by performing conjugation assay using E. coli (ThermoFisher™, USA) as recipient cells.20 Initially, LB media and LB broth (HIMEDIA®, India) were prepared, autoclaved, and pour into two different sets of plates and culture tubes. In the first set of plates and tubes, only LB media and LB broth were poured without adding anything and labeled. In other sets of culture tubes, LB broth was supplemented with DAP (diaminopimelic acid, 0.3 mM), ceftriaxone (30 µg/mL) and meropenem (10 µg/mL) and labeled. A fresh K. pneumoniae suspension was prepared in LB broth having the antibiotics by incubating over nightly. Fresh recipients E. coli suspension was prepared in plain LB broth by incubating over nightly. In a tube, 50+50 µL of both cells were mixed by repeated pipetting to make a ratio of 1:1 and 100 µL of mixed cell suspension was poured on LB agar plates containing meropenem (10 µg/mL) and labeled as “conjugation plate”, kept at 37 °C for 24 hours. Moreover, PBS suspension of the bacterial colony was used for further confirmation of the transconjugants via PCR using specific primers against the blaNDM-1 gene.
Statistical Analysis
The differences in the percentage/proportions of K. pneumonia bacteria isolated from different sources were tested by using “prop. Test” function in R software. This test is used for testing the null that the proportions in each group are the same (R Core Team, 2017).
Results
Distribution of K. pneumoniae According to the Respective Sample Source
The findings of the study revealed that the overall prevalence of confirmed carbapenem-resistant K. pneumoniae was 9.4% (13/138). The highest prevalence of 10.7% (7/65) was recorded from abattoir/wastewater samples followed by 10% (4/40) prevalence from milk and fecal samples While 6 % (2/33) prevalence was found in veterinary sludge samples (Table 1). Statistically, the results from Pearson’s chi-squared test statistic (using prop. test in R software) showed that the difference between proportions or percentage of occurrence of K. pneumoniae isolated from seven different sources (shown in Table 1) is non-significant (p = 0.96, Chi-squared statistics = 1.35, df = 7).
Resistance Profile of the Isolates
For the 13 carbapenem-resistant isolates, the disc diffusion assay showed that all the isolates 100% (13/13) of K. pneumoniae were resistant to all the generations of cephalosporins as well as to meropenem. The resistance to ciprofloxacin, tetracyclines, and trimethoprim-sulfamethoxazole were 76.9% and 46.1% and 38.46% respectively as shown in Table 3.
 |
Table 3 Distribution of MICs of Carbapenem-Resistant Klebsiella pneumoniae (KP) Isolates Belonging to Various Multilocus Sequences Typing (MLST) |
Distribution of ARGs
After phenotypic characterization, all the carbapenem-resistant K. pneumoniae isolates were subjected to PCR for the detection of various ARGs. Among various ESBL genes blaTEM was detected in all (100%; 13/13) the isolates followed by blaSHV (92.30%; 12/13) and blaCTXM-1 (84.6%; 11/13). Among various MBL genes blaNDM-1 was detected in 92.30% (12/13) isolates followed by blaOXA-48 84.61% (11/13), while blaKPC was detected in 01 (7.69%) isolate only. Whereas, blaIMP, blaVIM, and blaGIM were not detected in any isolate of the study. Other detected ARGs include, qnrA 0%, qnrB 53.84% (7/13) and qnrS 46.1% (6/13), tetA 30.7% (4/13), tetB 23% (3/13), sul1 and sul2 30.7% (4/13). Details of the ARGs distribution in K. pneumoniae isolates along their respective PCR product size were given in Table 2. The distribution of different Beta-lactamases genes among carbapenem resistant K. pneumoniae isolates is shown in Table 4.
 |
Table 4 Co-Existence of Different β-Lactamases and Metallo-β-Lactamases Encoding Genes Among Carbapenem-Resistant Klebsiella pneumoniae (KP) Isolates |
Confirmation of Transconjugants
Bacterial colonies that appeared on LB agar plates supplemented with meropenem (10 µg/mL) were considered as transconjugants. Additionally, blaNDM-1 was detected in all the transconjugants subjected to colony PCR. All the transconjugants displayed a strong resistance pattern against meropenem, as all the transconjugants showed a MIC of 128 µg/mL, which is significantly higher than the interpretative breakpoint given by CLSI.
Prevalent Sequence Types of K. pneumoniae in Veterinary Sample Sources
MLST results revealed that overall, three different STs of K. pneumoniae were detected in the study which includes ST 29, ST11 and ST 258. Among these STs, ST 29 was found in 53.84% (7/13) isolates followed by ST11 which was found in 38.46% (5/13) isolates whereas, single isolate (7.69%; 1/13) of the study showed the allelic profile of ST258. It was found that ST29 was the most prevalent ST in abattoir wastewater, as all the isolates 100% (3/3) from abattoir wastewater displayed the allelic profile of ST29. While both isolates 100% (2/2) from fecal samples belong to ST11. The single isolate of K. pneumoniae which showed the allelic profile of ST258 was isolated from poultry sludge (Table 3).
Discussion
During the last few years, gram-negative bacteria displayed a significant increase in the resistance against beta-lactam antibiotics, because of different plasmid-mediated ESBL genes, present in the Enterobacteriaceae family especially in E. coli and K. pneumoniae. The emergence of resistant K. pneumoniae due to the abuse of antibiotics in livestock farms and veterinary settings is a serious public as well as livestock health concern; it may ultimately be disseminated to the human via various environmental niches.21
Pakistan is an agriculture-based country, 70% of people directly or indirectly are involved with agriculture particularly livestock farming. Intense modern livestock farming practices force them to use antibiotics as feed additives, which ultimately associated with the development and dissemination of antibiotic-resistant superbugs. In this context, the prevalence of carbapenem-resistant K. pneumoniae (CRKP) has been well reported from various regions of the world.3,7 However, data regarding the prevalence of CRKP from Pakistan is very limited. Here, in the present study, we demonstrated the prevalence of carbapenem-resistant K. pneumoniae in veterinary settings and waste from Pakistan.
Findings revealed that overall 9.4% (13/138) prevalence of CRKP was observed in various samples from veterinary settings and waste. The prevalence was higher in wastewater samples compared to the other samples source (Table 1). Yet no such study has reported the prevalence of CRKP in veterinary settings and waste from Pakistan. However, the prevalence of CRKP has been well documented in Veterinary settings and waste in the world.4,22 Livestock farms have also been recognized as a reservoir of ARGs and their dissemination in Jiangsu Province, China.23 Similarly, some studies from India also reported that animal milk and meat, etc. are the potential source of ESBL producing K. pneumoniae.24,25 In this study, we also have isolated CRKP from milk and slaughterhouse waste. The possible explanation of this high prevalence is that antibiotics are aberrantly used in veterinary practice here in Pakistan and as a growth promoter in food animals, which ultimately steer the evolution of antibiotic-resistant bacterial strains. Several studies have documented that food-producing animals are the possible source or reservoir for the dissemination of resistant bacterial strains or ARGs to humans.26,27 While, another possible reason for the high prevalence of CRKP in milk may be due to mastitis, as bovine mastitis is a substantial factor for the growth and colonization of bacterial pathogens including K. pneumoniae.26 In the past, a study reported the detection of ESBL producing K. pneumoniae in the milk of healthy cows.28
Different ARGs (Table 2) have been detected in the isolates of the study. Particularly, a total of 12 (92.3%) isolates were positive for the blaNDM-1 gene and 11 (84.6%) isolates were positive the blaOXA-48. The occurrence of the blaNDM-1 gene in food-producing animals has already been reported form different parts of the world29 but no such report is published from Pakistan. Additionally, several studies stated that usually blaNDM-1 gene is located on mobile a genetic element that carry few additional ARGs as well which helps in the dissemination of blaNDM-1.30 The Indian subcontinent is recognized, as a reservoir of blaNDM-1 harboring Enterobacteriaceae. Moreover, many studies have also demonstrated the distribution of carbapenem-resistant Enterobacteriaceae in livestock from all across the globe.31 On the other hand, the incidence of blaNDM-1 harboring K. pneumoniae has only been well documented in clinical and hospital settings from Pakistan.32,33 It is very tough to compare our results with local findings, as according to our best knowledge yet no study has reported the prevalence of blaNDM-1 harboring K. pneumoniae in veterinary settings and waste from Pakistan.
All the CRKP isolates were subjected to conjugation assay to check the transferability of the blaNDM-1 gene. As expected, it was observed that blaNDM-1 was transferable to the donor E. coli cells, indicating that irrespective of the ST, all the blaNDM-1 was located on conjugative plasmids and a significant source of horizontal gene transfer (HGT). The range of conjugation efficiency was 1.0 x 10−6 and 3.5 x 10−6 per transconjugant which also suggested the probability of HGT. Different sample niches especially veterinary sludge and wastewater may have high bacterial density and there would be a significant chance for horizontal gene transfer of blaNDM-1 to various bacterial strains present in these sources. The same recommendations have been made in the past that resistance genes may be disseminated through bacterial strains via mobile genetic elements and plasmids. Additionally, detailed molecular studies have been reported that K. pneumoniae strains may disseminate different ARGs particularly blaNDM-1 and blaOXA-48 via HGT.6,34
To know the sequence types of the isolates MLST was performed. During MLST three discrete STs of K. pneumoniae were observed which include ST29 (46.1%), ST11 (38.5%) and ST 258 (7.7%). In this study, ST29 in a leading sequence type which is quite interesting. The same results have been reported from Saudi Arabia, where they found that ST29 was the predominant ST of carbapenem-resistant K. pneumoniae in Riyadh.35 Incidence of K. pneumoniae ST29 in veterinary products has been reported from various regions, a study conducted in Ghana reported the occurrence of ESBL producing K. pneumoniae ST29 in local and imported poultry meat.36,37 Several studies have described the transmission of ESBL harboring Enterobacteriaceae from food animals and their products to humans. Detection of similar clones of ESBL- harboring E. coli and K. pneumoniae in animals and humans provides indirect proof of such cross species-transmission.38 In this study, we have reported ST11 from veterinary settings of Pakistan for the very first time, before that ST11 has only been documented in clinical and hospital settings in Pakistan.33 Whereas, ST11 has been recognized as the most prevalent ST in Asia particularly in China and Taiwan.37,39 Detection of K. pneumoniae ST258 in the present study is also worrisome, as ST258 is one of the most detected STs of K. pneumoniae in the world.40,41 Few factors contribute substantially to the global expansion of ST258 are virulence genes, type IV secretion system, type IV pilus, and type-III restriction-modification system.42
Conclusion
To best of our knowledge, this is the first study from Pakistan which demonstrated that veterinary settings and waste are the potential sources of carbapenem-resistant K. pneumoniae. In Pakistan, sanitation facilities are inadequate, which is a considerable risk factor associated with the dissemination of CRKP to the community. Strong surveillance and monitoring policy are required to estimate the exact burden of this public and livestock health menace which would be beneficial to curtail this health concern.
Data Sharing Statement
The aggregate data supporting findings contained within this manuscript will be shared upon request submitted to the corresponding author. Identifying patient data will not be shared.
Acknowledgments
We are grateful to the HEC Pakistan for IRSIP fellowship awarded to Dr. Tamoor Hamid Chaudhry at the Paul G. Allen School for Global Animal Health, Washington State University, Washington, USA.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Disclosure
The authors have no conflicts of interest to declare for this work.
References
1. Aslam B, Wang W, Arshad MI, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645–1658. doi:10.2147/IDR.S173867
2. Baloch Z, Aslam B, Muzammil S, Khurshid M, Rasool MH, Ma K. Selection inversion: a probable tool against antibiotic resistance. Infect Drug Resist. 2018;11:1903–1905. doi:10.2147/IDR.S176759
3. Aslam B, Chaudhry TH, Arshad MI, et al. The first bla KPC harboring Klebsiella pneumoniae ST258 strain isolated in Pakistan. Microbial Drug Resistance. 2020;26:783–786. doi:10.1089/mdr.2019.0420
4. Hiroi M, Yamazaki F, Harada T, et al. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in food-producing animals. J Vet Med Sci. 2011;1109290647.
5. Geser N, Stephan R, Hachler H. Occurrence and characteristics of extended-spectrum beta-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet Res. 2012;8:21. doi:10.1186/1746-6148-8-21
6. Hudson CM, Bent ZW, Meagher RJ, Williams KP. Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. PLoS One. 2014;9(6):e99209. doi:10.1371/journal.pone.0099209
7. Hayat S, Siddique MH, Aslam B, et al. Extended-spectrum-β-lactamase producing multidrug resistant klebsiella pneumoniae isolates from pediatrics. Pak J Zool. 2019;51:4. doi:10.17582/journal.pjz/2019.51.4.1251.1257
8. Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–5054. doi:10.1128/AAC.00774-09
9. Hammerum AM, Toleman MA, Hansen F, et al. Global spread of New Delhi metallo-β-lactamase 1. Lancet Infect Dis. 2010;10(12):829–830. doi:10.1016/S1473-3099(10)70276-0
10. Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(suppl_1):S28S36. doi:10.1093/infdis/jiw282
11. Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873–5884. doi:10.1128/AAC.01019-15
12. Khurshid M, Rasool MH, Ashfaq UA, Aslam B, Waseem M. Emergence of ISAba1 harboring carbapenem-resistant Acinetobacter baumannii isolates in Pakistan. Future Microbiol. 2017;12:1261–1269. doi:10.2217/fmb-2017-0080
13. Khurshid M, Rasool MH, Ashfaq UA, et al. Dissemination of bla(OXA-23) harboring carbapenem-resistant acinetobacter baumannii clones in Pakistan. J Global Antimicrobial Resistance. 2020;21:357–362. doi:10.1016/j.jgar.2020.01.001
14. Khurshid M, Rasool MH, Siddique MH, et al. Molecular mechanisms of antibiotic co-resistance among carbapenem resistant Acinetobacter baumannii. J Infect Dev Ctries. 2019;13(10):899–905. doi:10.3855/jidc.11410
15. Bonomo RA. New Delhi metallo-β-lactamase and multidrug resistance: a global SOS? Clin Infectious Diseases. 2011;52(4):485–487. doi:10.1093/cid/ciq179
16. Mohsin M, Van Boeckel TP, Saleemi MK, et al. Excessive use of medically important antimicrobials in food animals in Pakistan: a five-year surveillance survey. Glob Health Action. 2019;12(sup1):1697541. doi:10.1080/16549716.2019.1697541
17. Chaudhry TH, Aslam B, Arshad MI, Nawaz Z, Waseem M. Occurrence of ESBL-producing Klebsiella pneumoniae in hospital settings and waste. Pak J Pharm Sci. 2019;32(2(Supplementary)):773–778.
18. In C. Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standards Institute; 2018.
19. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178–4182. doi:10.1128/JCM.43.8.4178-4182.2005
20. Ferrieres L, Hémery G, Nham T, et al. Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery. J Bacteriol. 2010;192(24):6418–6427. doi:10.1128/JB.00621-10
21. Lyhs U, Ikonen I, Pohjanvirta T, Raninen K, Perko-Mäkelä P, Pelkonen S. Extraintestinal pathogenic Escherichia coli in poultry meat products on the Finnish retail market. Acta Vet Scand. 2012;54(1):64. doi:10.1186/1751-0147-54-64
22. Ohnishi M, Okatani A, Esaki H, et al. Herd prevalence of Enterobacteriaceae producing CTX‐M‐type and CMY‐2 β‐lactamases among Japanese dairy farms. J Appl Microbiol. 2013;115(1):282–289. doi:10.1111/jam.12211
23. Chen B, Hao L, Guo X, Wang N, Ye B. Prevalence of antibiotic resistance genes of wastewater and surface water in livestock farms of Jiangsu Province, China. Environ Sci Pollut Res. 2015;22(18):13950–13959. doi:10.1007/s11356-015-4636-y
24. Koovapra S, Bandyopadhyay S, Das G, et al. Molecular signature of extended spectrum β-lactamase producing Klebsiella pneumoniae isolated from bovine milk in eastern and north-eastern India. Infect Genet Evol. 2016;44:395–402. doi:10.1016/j.meegid.2016.07.032
25. Samanta I, Joardar SN, Mahanti A, Bandyopadhyay S, Sar TK, Dutta TK. Approaches to characterize extended spectrum beta-lactamase/beta-lactamase producing Escherichia coli in healthy organized vis-a-vis backyard farmed pigs in India. Infect Genet Evol. 2015;36:224–230. doi:10.1016/j.meegid.2015.09.021
26. Saishu N, Ozaki H, Murase T. CTX-M-type extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolated from cases of bovine mastitis in Japan. J Vet Med Sci. 2014;76(8):1153–1156. doi:10.1292/jvms.13-0120
27. Timofte D, Maciuca IE, Evans NJ, et al. Detection and molecular characterization of Escherichia coli CTX-M-15 and Klebsiella pneumoniae SHV-12 β-lactamases from bovine mastitis isolates in the United Kingdom. Antimicrob Agents Chemother. 2014;58(2):789–794. doi:10.1128/AAC.00752-13
28. Hammad AM, Ahmed AM, Ishida Y, Shimamoto T. First characterization and emergence of SHV-60 in raw milk of a healthy cow in Japan. J Vet Med Sci. 2008;70(11):1269–1272. doi:10.1292/jvms.70.1269
29. Zhang W-J, Lu Z, Schwarz S, et al. Complete sequence of the bla NDM-1-carrying plasmid pNDM-AB from Acinetobacter baumannii of food animal origin. J Antimicrob Chemother. 2013;68(7):1681–1682. doi:10.1093/jac/dkt066
30. Villa L, Poirel L, Nordmann P, Carta C, Carattoli A. Complete sequencing of an IncH plasmid carrying the bla NDM-1, bla CTX-M-15 and qnrB1 genes. J Antimicrob Chemother. 2012;67(7):1645–1650. doi:10.1093/jac/dks114
31. Fischer J, Rodríguez I, Schmoger S, et al. Escherichia coli producing VIM-1 carbapenemase isolated on a pig farm. J Antimicrob Chemother. 2012;67(7):1793–1795. doi:10.1093/jac/dks108
32. Nahid F, Khan AA, Rehman S, Zahra R. Prevalence of metallo-β-lactamase NDM-1-producing multi-drug resistant bacteria at two Pakistani hospitals and implications for public health. J Infect Public Heal. 2013;6(6):487–493. doi:10.1016/j.jiph.2013.06.006
33. Qamar MU, Saleem S, Toleman MA, et al. In vitro and in vivo activity of Manuka honey against NDM-1-producing Klebsiella pneumoniae ST11. Future Microbiol. 2018;13(1):13–26. doi:10.2217/fmb-2017-0119
34. Kelly BG, Vespermann A, Bolton DJ. Gene transfer events and their occurrence in selected environments. Food Chem Toxicol. 2009;47(5):978–983. doi:10.1016/j.fct.2008.06.012
35. Uz Zaman T, Aldrees M, Al Johani SM, Alrodayyan M, Aldughashem FA, Balkhy HH. Multi-drug carbapenem-resistant Klebsiella pneumoniae infection carrying the OXA-48 gene and showing variations in outer membrane protein 36 causing an outbreak in a tertiary care hospital in Riyadh, Saudi Arabia. Int j Infectious Diseases. 2014;28:186–192. doi:10.1016/j.ijid.2014.05.021
36. Eibach D, Dekker D, Gyau Boahen K, et al. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in local and imported poultry meat in Ghana. Vet Microbiol. 2018;217:7–12. doi:10.1016/j.vetmic.2018.02.023
37. Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66(2):307–312. doi:10.1093/jac/dkq431
38. Leverstein‐van Hall M, Dierikx C, Cohen Stuart J, et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infection. 2011;17(6):873–880. doi:10.1111/j.1469-0691.2011.03497.x
39. Tseng IL, Liu YM, Wang SJ, et al. Emergence of carbapenemase producing klebsiella pneumonia and spread of KPC-2 and KPC-17 in Taiwan: a nationwide study from 2011 to 2013. PLoS One. 2015;10(9):e0138471. doi:10.1371/journal.pone.0138471
40. Bowers JR, Kitchel B, Driebe EM, et al. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae Pandemic. PLoS One. 2015;10(7):e0133727. doi:10.1371/journal.pone.0133727
41. Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22(12):686–696. doi:10.1016/j.tim.2014.09.003
42. Chen L, Mathema B, Pitout JD, DeLeo FR, Kreiswirth BN. Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. MBio. 2014;5(3):e0135501314. doi:10.1128/mBio.01355-14
43. Kurupati P, Chow C, Kumarasinghe G, Poh CL. Rapid detection of Klebsiella pneumoniae from blood culture bottles by real-time PCR. J Clin Microbiol. 2004;42:1337–1340. doi:10.1128/JCM.42.3.1337-1340.2004
44. Schlesinger J, Navon-Venezia S, Chmelnitsky I, et al. Extended-spectrum beta-lactamases among Enterobacter isolates obtained in Tel Aviv, Israel. Antimicrob Agents Chemother. 2005;49:1150–1156. doi:10.1128/AAC.49.3.1150-1156.2005
45. Krishnamurthy V, Vijaykumar G, Kumar S, Prashanth H, Prakash R, Nagaraj E. Phenotypic and genotypic methods for detection of extended spectrum β lactamase producing Escherichia coli and Klebsiella pneumoniae isolated from ventilator associated pneumonia. J Clin Diagn Res. 2013;7:1975.
46. Gootz TD, Lescoe MK, Dib-Hajj F, et al. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob Agents Chemother. 2009;53:1998–2004. doi:10.1128/AAC.01355-08
47. Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-β-lactamases. J Antimicrob Chemother. 2006;59:321–322. doi:10.1093/jac/dkl481
48. Huang T-W, Chen T-L, Chen Y-T, et al. Copy number change of the NDM-1 sequence in a multidrug-resistant Klebsiella pneumoniae clinical isolate. PLoS One. 2013;8:e62774. doi:10.1371/journal.pone.0062774
49. Poirel L, Castanheira M, Carrër A, et al. OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 2011;55:2546–2551. doi:10.1128/AAC.00022-11
50. Wang A, Yang Y, Lu Q, et al. Presence of qnr gene in Escherichia coli and Klebsiella pneumoniae resistant to ciprofloxacin isolated from pediatric patients in China. BMC Infect Dis. 2008;8:68. doi:10.1186/1471-2334-8-68
51. Bokaeian M, Saeidi S, Shahi Z, Kadaei V. TetA and tetB Genes in Klebsiella pneumoniae isolated from clinical samples. Gene, Cell and Tissue 1. 2014;1. doi:10.17795/gct-18152
52. Frank T, Gautier V, Talarmin A, Bercion R, Arlet G. Characterization of sulphonamide resistance genes and class 1 integron gene cassettes in Enterobacteriaceae, Central African Republic (CAR). J Antimicrob Chemother. 2007;59:742–745. doi:10.1093/jac/dkl538
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
