Back to Journals » Journal of Inflammation Research » Volume 15
EGF Protects Epithelial Cells from Barrier Damage in Chronic Rhinosinusitis with Nasal Polyps
Authors Chen L, Liu Q, Liu Z, Li H, Liu X, Yu H
Received 22 October 2021
Accepted for publication 24 December 2021
Published 19 January 2022 Volume 2022:15 Pages 439—450
DOI https://doi.org/10.2147/JIR.S345664
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Ning Quan
Le Chen,1,2 Quan Liu,1,2 Zhuofu Liu,1,2 Han Li,1,2 Xiang Liu,1,2 Hongmeng Yu1,2
1Department of Otolaryngology, Eye & ENT Hospital, Fudan University, Shanghai, 200031, People’s Republic of China; 2Research Units of New Technologies of Endoscopic Surgery in Skull Base Tumour, Chinese Academy of Medical Sciences, 2018RU003, Shanghai, People’s Republlc of China
Correspondence: Hongmeng Yu; Xiang Liu Email [email protected]; [email protected]
Background: The aim of this study is to investigate the potential key genes related to Chronic rhinosinusitis with nasal polyps (CRSwNP).
Methods: Datasets GSE36830 and GSE72713 were obtained from Gene Expression Omnibus. Dataset GSE36830 was used to identify differentially expressed genes in CRSwNP patients. GO, KEGG analysis, and PPI network analysis were applied to further investigate the function of DEGs in CRSwNP. GSEA was also performed to explore the mechanisms of DEGs. Dataset GSE72713 was applied to validate the key gene. Moreover, to detect the expression of target gene, nasal polyp tissues and middle turbinate specimens were collected from CRSwNP patients (n = 20) and controls (n = 20), respectively. RT-PCR, Western blot, and immunofluorescence staining were applied. HE and AB-PAS staining were used to assess the infiltration of inflammatory cells. The proliferation and migration ability of human nasal epithelial cells (HNEpCs) were tested via Cell Counting kit-8, wound healing assay and Transwell migration assay. Air–liquid interface was used to culture primary human nasal epithelial cells (pHNECs) from health controls and nasal polyp tissues of CRSwNP patients.
Results: A total of 1035 DEGs were identified, and 661 genes were up-regulated and 374 genes were down-regulated. According to PPI network analysis, the top 10 scored genes were identified. Among them, only EGF was down-regulated in CRSwNP. Meanwhile, GSEA result shows that EGF is significantly enriched in WNT activated receptor activity. CCK-8, wound healing assay, and transwell migration assay indicated that recombinant human EGF can promote the proliferation and migration of HNEpCs in vitro. Immunofluorescence staining shows that rhEGF can increase the expression of ZO-1 in pHNECs from nasal polyp tissues.
Conclusion: Bioinformatics analysis and in vitro experiments were used to explore the pathogenesis of CRSwNP, and the results showed that EGF may play an important role in the protection of nasal epithelial barrier.
Keywords: chronic rhinosinusitis with nasal polyps, bioinformatics analysis, GSEA, EGF, epithelial barrier
Introduction
Chronic rhinosinusitis (CRS) is a common chronic nasal mucosa inflammation disease, which affects approximately 7% to 27% of adult population.1–3 It is a group of heterogeneous diseases with common symptoms but different pathogenesis.4,5 Based on the presence or absence of nasal polyps (NP), CRS has been divided into CRS with NP (CRSwNP) and CRS without NP (CRSsNP).6 CRSwNP is characterized by the nasal obstruction and inflammation of the nasal mucosa, which seriously reduces the quality of life of patients.7–10 The etiologies of CRSwNP have been investigated worldwide and extraordinary progress has been made.7,11,12 The nasal epithelium is the physical and immune barrier, and its dysfunction is an outstanding feature of CRSwNP.13–18 Epithelial cells play an important role in innate immune, and it can produce protective proteins and resist microbial invasion.19–22 Meng et al revealed that the expression levels of ZO-1, E-cadherin, and occludin were down-regulated in mature polyps, and epithelial damage was most obviously in early stage.23 Meanwhile, several studies have reported the decreased expression of ZO-1, E-cadherin and occludin in polyps.23–28 It is currently believed that the regeneration and repairment of epithelium are changed in patients with CRSwNP, and the reduced barrier function of epithelium is a potent contributing factor to type 2 immunity.29–32 However, the mechanisms underlying CRSwNP are not fully defined. Therefore, to develop a novel treatment for CRSwNP, further investigation is needed.
High throughput sequencing is an increasingly important field. It has been widely applied in biological research. In the present study, the differentially expressed genes (DEGs) of nasal epithelium between CRSwNP patients and healthy individuals were analyzed based on the microarray datasets downloaded from the Gene Expression Omnibus (GEO) database. In the dataset GSE36830, Seshadri et al collected nasal tissues from patients with skull base tumor and patients with CRS.11 Gene Ontology (GO) process, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, protein–protein interaction (PPI) networks, and Gene set enrichment analysis (GSEA) were performed to identify the key genes and pathways associated with CRSwNP in the dataset GSE36830. Dataset GSE72713 was downloaded and applied to validate the identified potential target gene in dataset GSE36830.33 Moreover, cell biology experiments were used to investigate the effects of target gene in CRSwNP. As far as we know, the present study is one of the first to integrate GSEA and biological experiments to explore the mechanism involved in CRSwNP.
Methods
Microarray Data Processing
Dataset GSE36830 acquired from the GEO database was used to identify the key genes related to CRS. The platform of GSE36830 dataset was GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array). This dataset included NP tissues from 6 subjects with CRSwNP, uncinate tissues (UT) from 6 subjects with CRSsNP and 6 control subjects. To validate the target gene, dataset GSE72713 was downloaded. In dataset GSE72713, the data of total RNA was extracted from NP tissues of 6 CRSwNP patients and sphenoid sinus mucosa of 3 normal controls.
Identification of DEGs
R software (version 3.5.2, United States) was used to normalize the raw microarray data of GSE36830. The “limma” package in R software was carried out to identify the DEGs between the NP tissues of CRSwNP patients and the health controls. In addition, the DEGs between the UT tissues of the CRSsNP patients and the NP tissues of CRSwNP patients, and the UT tissues of the CRSwNP patients and the NP tissues of CRSwNP patients were identified. |log FC| (fold change) >1 and p < 0.05 were considered as statistical significance.
Construction of the PPI Networks
The Search Tool for the Retrieval of Interacting Genes (STRING) platform was used to construct the PPI networks of DEGs in different groups. The value of the correlation between proteins was set to greater than 0.4. Cytohubba, a plug-in of Cytoscape software (version 3.7.1, UAS) was applied to identify the top 10 key genes of connectivity of DEGs in different groups.
GO and KEGG Analyses
Database for Annotation, Visualization, and Integrated Discovery (DAVID) is usually used for functional analysis of genes and proteins. To further investigate the function of DEGs between the NP tissues of CRSwNP patients and the health controls, DAVID was applied to carry out the GO process and KEGG pathway analyses. False discovery rate (FDR) <0.05 was set as the cutoff point.
Gene Sets Enrichment Analysis (GSEA)
To further investigate the pathogenesis of CRSwNP, GO analysis of all genes of the NP tissues of CRSwNP and the health controls in GSE36830 was carried out by GSEA software (version 4.0.3, USA). This analysis can explore the major pathway that the target genes involved in.
Patients and Specimens
The tissue samples were obtained from patients in the Department of Otolaryngology-Head and Neck Surgery, Eye, Ear, Nose, and Throat (Eye & ENT) Hospital of Fudan University. The normal control middle turbinate tissues were obtained from the patients (n = 20) who underwent skull base tumor excision or optic nerve decompression operation and had no history of upper airway inflammatory diseases. NP tissues were obtained from CRSwNP patients (n = 20) who underwent functional endoscopic sinus surgery. The exclusion criteria were as follows: age <18 or >80 years, a diagnosis of cystic fibrosis, immunodeficiency, aspirin intolerance, autoimmune disease, Churg-Strauss syndrome, asthma, or fungal sinusitis. This study complies with the Declaration of Helsinki and was approved by the Eye & ENT Hospital and all subjects gave informed consent.
Extraction of Total RNA and Quantitative RT-PCR
For extraction of total RNA, NP tissues and middle turbinate tissues were homogenized with 1 mL Trizol Reagent according to the guidelines. The complementary DNA (cDNA) template was synthesized with PrimeScript RT Master mix (TaKaRa, Japan). Then, the mRNA expression of the target gene was analyzed using an RT-PCR Sequence Detection System (ABI, USA) with SYBR Green chemistry (TaKaRa, Japan). The specific RT-PCR primer for EGF: forward 5ʹ-GAAGCATTGGACAAGTATGCAT −3ʹ, reverse 5ʹ-CAGCTTCTGAGTCCTGTAGTAG-3ʹ. The relative expression level of the target gene was analyzed using the 2−ΔΔCT method.34
Western Blot
Protein samples (20μg) obtained from frozen NP tissues and middle turbinate tissues were resolved by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride membranes (PVDF) (Millipore, USA). Then, it was incubated with anti-EGF antibody (1:700, Santa, USA) according to manufacturer’s protocol. The expression of β-actin (1:2000, Abcam, UK) was detected as an internal control. Densitometric analysis of the data was performed by Image J software (NIH, USA).
Histological Staining
To detect the infiltration of eosinophils and the hyperplasia of the mucous cells in the NP tissues and health controls, hematoxylin and eosin (HE) and Alcian blue-periodic acid Schiff (AB-PAS) staining were applied, as previously published.35–39 Slices were examined under microscope, and the number of eosinophils and mucous cells in different stainings were counted in 5 random high-power fields.
Immunofluorescence Staining
Briefly, slices were incubated with anti-tryptase antibody (1:100, Santa, USA) and anti-EGF antibody (1:700, Santa, USA) at 4°C overnight, and then incubated with the secondary antibody at room temperature for 2h.40 Immunofluorescence samples were observed and photographed with an inverted microscope (Leica, Germany). Image J software (NIH, USA) was used to analyze the mean fluorescence intensity.
Cell Culture
Human nasal epithelial cell (HNEpC) line was obtained from American Type Culture Collection (ATCC, USA). HNEpCs were cultured in RPMI1640 (Gibco, USA), supplemented with 10% fetal bovine serum (Gibco, USA) and 1% penicillin–streptomycin (Gibco, USA). Cells were cultured at 37°C with 5% CO2 atmosphere.
Cell Proliferation Assay
Approximately 2 × 103 HNEpCs were cultured on 96-well culture plates in 100 μL of RPMI1640 medium with 10% FBS and 1% penicillin-streptomycin. HNEpCs were cultured for 1~4 days at 37°C with 5% CO2 atmosphere. For EGF treatment group, cells were cultured with the above medium containing 100 ng/mL recombinant human EGF (rhEGF, PeproTech, USA). The proliferation was detected by Cell Counting Kit-8 (CCK-8) assay kit (Dojindo, Japan) according to the manufacturer’s instructions. Briefly, 10 μL of CCK-8 solution was added to each well for 1 h incubation, and the absorbance of the test wells was measured by using a microplate reader at 450 nm.
Wound Healing and Transwell Migration Assays
HNEpCs were seeded into 6-well culture plates. To form an artificial wound, cell monolayers were scraped off with a 200 μL pipette tip. After scratching, to remove the detached cells and residual serum, the wells were washed three times with PBS and then serum-free 1640 medium was added. For EGF treatment group, cells were cultured with 1640 medium containing 50 ng/mL and 100 ng/mL rhEGF. The wound healing of HNEpCs was imaged with an inverted microscope (Leica, Germany) at 0 h, 24 h and 48 h.
The migration of HNEpCs was investigated in Transwell chambers (8-μm pore filters, Corning, USA). Approximately 1 × 104 cells suspended in 100 μL serum-free 1640 medium were placed in the upper chamber, and 600 μL 1640 medium which contains 10% FBS or 10% FBS and 50, 100ng/mL rhEGF was added to the lower chamber, respectively. After 24 h and 48 h, the cells on the lower surface of the chamber were fixed, stained, and counted.
Primary Human Epithelial Cell Culture and rhEGF Stimulation
Primary human epithelial cells (pHNECs) were isolated from the above nasal tissues. Briefly, nasal samples were immediately immersed in DPBS (Ca2+/Mg2+-free) supplemented with 1% penicillin, streptomycin, and amphotericin B (all reagents from Gibco). After washed three times, nasal tissues were digested with 0.1% Protease XIV (Sigma-Aldrich) overnight at 4°C. After filtering, all cells were immersed in DMEM/F12 media (Gibco, USA) which contains 10% FBS and 1% ITS for 2 h at 37 °C with 5% CO2 atmosphere. Next, cells in supernatant media were cultured in PneumaCult-Ex Medium (StemCell, Canada). In addition, cells from NP tissues were exposed to 100 ng/mL rhEGF for 48 h. After reaching about 70%~80% confluence, cells were digested with Accutase (StemCell, Canada) for about 5 min, and then seeded on Transwell chamber (0.4 μm pore size; Corning, USA). To induce cell differentiation at ALI culture, the medium at the basal side of Transwell was replaced by PneumaCult-ALI Medium kit (StemCell, Canada) and the medium at the apical side was removed. PHNECs were fully mature after cultured for 21 days in ALI.
To identify the ALI cultured cells, HE, AB-PAS and immunofluorescence staining were applied. The expression of tubulin, p63, and mucin5AC (the marker of ciliated cell, basal cell and mucous cell) were detected by immunofluorescence.41 The slices were photographed with an inverted microscope (Leica, Germany). The systematic workflow of the study is shown in Figure 1.
 |
Figure 1 Workflow diagram of data collection and analysis. |
Statistical Analysis
All experimental data were presented as means ± SD. Comparisons were calculated using one-way ANOVA or Student’s t-test. All statistical analyses were performed using GraphPad prism 8.0 software. P value < 0.05 was considered statistically significant.
Results
Identification of DEGs and Key Gene
In dataset GSE36830, a total of 1035 DEGs were identified between the NP tissues and health controls by R software (|logFC| >1, p < 0.05), and the top 50 DEGs were shown in the heatmap (Figure 2A). Among them, 661 genes were up-regulated and 374 genes were down-regulated (Figure 2B). The PPI networks of DEGs were constructed based on the STRING database, and the cytohubba plug-in of Cytoscape was applied to identify the top 10 key genes (TYROBP, IL10, CD86, ITGB2, SPI1, EGF, LILRB2, CSF1R, CCR7, and FCGR2B) (Figure 2C and D). Only the expression of EGF was down-regulated in NP group among the top 10 key genes (Supplemental Figure 1). In addition, the DEGs and identified key genes between the UT of CRSsNP and NP of CRSwNP group, and those between the UT of CRSwNP and NP of CRSwNP group are shown in Supplemental Figure 2.
GO and KEGG Analyses
DAVID online tool was used for functional analysis of the 1035 DEGs between the NP tissues and health controls. GO functional enrichment analysis showed that the DEGs were mainly significantly enriched in immune response, extracellular region, and integral component of plasma membrane (top 3) (Figure 2E). Meanwhile, KEGG analysis showed that they were mainly significantly enriched in Staphylococcus aureus infection, Cytokine–cytokine receptor interaction, and Hematopoietic cell lineage (top 3) (Figure 2F).
GO Enrichment Analysis of All Detected Genes
According to GSEA, EGF was highly enriched in “Wnt-activated receptor activity” (Figure 2G and H). The enrichment score (ES) = 0.57, and the normalized enrichment score (NES) = 1.65 (p<0.05).
The Validation of EGF in GSE72713 Dataset and Patient Tissues
The expression of EGF was decreased in NP tissues in GSE72713 dataset (Figure 3A). However, there were only 3 normal controls and 6 CRSwNP patients in GSE72713. To make the results more credible, patient tissues were collected in our hospital. The RT-PCR and WB results revealed that the expression levels of EGF mRNA and protein were significantly down-regulated in NP tissues compared with the normal controls (p<0.01) (Figure 3B–D).
Histological Staining Results
The HE, AB-PAS, and immunofluorescence staining showed that the number of eosinophils, mucous cells, and mast cells were significantly increased in NP tissues (Figure 3E–G). In addition, the results of immunofluorescence staining also revealed that the expression levels of EGF and ZO-1 were down-regulated in NP tissues (Figure 3H and I). EGF protein was mainly expressed in the ciliated cells and basal cells (Figure 3J).
EGF Regulates Proliferation and Migration of HNEpCs
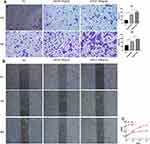
To investigate the role of EGF in the proliferation and migration of HNEpCs, cells were treated with rhEGF. The results of wound healing, transwell migration, and CCK-8 assays showed that rhEGF can promote proliferation and migration of HNEpCs (Figure 4A–C).
EGF Regulates the Expression of ZO-1
To further investigate the role of EGF in CRS, NP tissues and health controls were obtained and pHNECs were cultured. Cytomorphology of pHNECs was observed under optical microscope (Figure 5A). Cells were cultured in ALI for about 21 days, then HE and AB-PAS staining were used to detect the differentiation. We can see the clear hair-like structures of ciliated cells and AB-PAS positive mucous cells (Figure 5B). Meanwhile, immunofluorescence staining revealed the positive reaction to p63, tubulin, and mucin5AC antibodies, which was the marker of ciliated cell, basal cell and mucous cell, respectively (Figure 5C). The expression of ZO-1 was down-regulated in pHNECs from NP tissues compared with pHNECs from health controls, while the expression of ZO-1 was up-regulated in pHNECs from NP tissues which treated with 100 ng/mL rhEGF (Figure 5D).
Discussion
In the present study, GO, KEGG and PPI analyses were applied to investigate key genes involved in the pathogenesis of CRSwNP. By GO and KEGG analyses, immune response and Staphylococcus aureus infection were found to be the most significantly enriched terms, respectively (Figure 2E and F). The heterogeneity and clinical manifestations of CRS were dependent on the selective expression of type 1, 2, or 3 immune responses.42–44 Staphylococcus aureus is more usually exists in patients with CRSwNP compared with CRSsNPs, and it can be found under the epithelial surface.45–49 Staphylococcus aureus exoproducts alter epithelial repair on pHNECs significantly, and it is more obvious in the patients with nasal polyps.50 Staphylococcus aureus has a key impact on the nasal epithelium barrier and immune system in patients with CRS.51–59 According to PPI analysis, the top 10 key genes were identified by Cytohubba, among them, only EGF was down-regulated in NP tissues (Supplemental Figure 1). EGF (epidermal growth factor) is a 160 kDa membrane glycoprotein. It is a ligand of EGF-receptor and plays important roles in cell differentiation, proliferation, and motility.60 Jacob et al indicated that EGF was significantly down-regulated in atopic dermatitis patients, and this low-expression state may impair anti-inflammatory immune response and proliferation in the skin.61–64 Duan et al revealed that EGF was localized within p63+ basal cells in health controls, and there was a down-regulation of EGF in NP tissues.65 While, Ding et al reported an elevated level of EGF in the sinus mucosa of CRSwNP patients.66 In the present study, to detect the expression level of EGF in NP tissues and health controls, RT-PCR, WB and immunofluorescence staining analyses were applied. The results showed that the expression levels of EGF mRNA and protein were significantly down-regulated in NP tissues (Figure 3B–D and H). Meanwhile, down-regulated expression of ZO-1 protein was detected in NP tissues (Figure 3I). The immunofluorescence staining results showed that EGF was localized in p63+ basal cells and ciliated cells (Figure 3J). As reported in previous study, EGF was expressed in ciliated and basal cells in bronchial epithelium.67 The decreased number of ciliated cells may be associated with the down-regulated expression of EGF in NP tissues. To further explore the effects of EGF in nasal epithelial cells, biological experiments were carried out. From the results of wound healing, Transwell migration, and CCK-8 assays, we can see that rhEGF can promote the migration and proliferation of HNEpCs in vitro (Figure 4A–C). Moreover, pHNECs were successfully cultured in ALI. The pHNECs from NP tissues have a lower expression of ZO-1 compared to those from health controls. However, the expression of ZO-1 was up-regulated by treating with 100ng/mL rhEGF (Figure 5D). As previous studies reported that there was a decrease of ZO-1 in NP tissues of CRS.17 Sheng et al revealed that EGF down-regulated the expression of ZO-1 in pancreatic cancer cells.68 However, Okuyama et al reported the ability of EGF to prevent acid-induced decrease of ZO-1 in Human esophageal epithelial cells (TE-1).69 The relationship of EGF and ZO-1 in CRS still needs further research.
In addition, according to GSEA, we found that EGF was most enriched in WNT-activated receptor activity pathway (Figure 2G and H). Wnt signaling was found to become re-expressed in inflammatory diseases, and it played a huge part in injury repair.70–79 Boscke et al reported that Wnt signaling was up-regulated in NP tissues and this activation can promote the release of cytokines in HNEpCs.80 Meanwhile, the activation of Wnt pathway in HNEpCs can lead to the abnormal epithelial morphology and the loss of cellular differentiation. However, Whyte et al reported that Wnt signaling is responsible for the regenerating response of injured tissues.81 EGF and Wnt signaling are involved in the proliferation of intestinal epithelial stem cells and skin epithelial stem cells.82–88 EGF can mediate the proliferation of mesenchymal stem cells via Wnt/β-catenin activation.89 As far as we know, there have been few relevant researches in CRS, and intensive study is needed to reveal the effects of EGF and Wnt signaling on nasal epithelium.
Conclusion
In conclusion, this study identified the key genes and pathways involved in CRSwNP by bioinformatics analysis, and the key gene was verified by patient samples. Meanwhile, we demonstrated the promotional effects of EGF on the proliferation and migration of HNEpCs in vitro, and EGF can lead to an increased expression of ZO-1 in pHNECs from NP tissues.
Data Sharing Statement
All datasets generated for this study are included in the article.
Ethics Statement
This study was reviewed and approved by the Eye & ENT Hospital.
Acknowledgments
This work was supported by the National Natural Science Foundation of China under Grant number 81700901; Shanghai Science and Technology Committee Foundation under Grant number 19411950600; the New Technologies of Endoscopic Surgery in Skull Base Tumor: CAMS Innovation Fund for Medical Sciences (CIFMS) under Grant number 2019-I2M-5-003.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work. All authors read and approved the final manuscript.
Disclosure
The authors declare that they have no competing interests.
References
1. Hastan D, Fokkens WJ, Bachert C, et al. Chronic rhinosinusitis in Europe–an underestimated disease. A GA(2)LEN study. Allergy. 2011;66:1216–1223. doi:10.1111/j.1398-9995.2011.02646.x
2. Shi JB, Fu QL, Zhang H, et al. Epidemiology of chronic rhinosinusitis: results from a cross-sectional survey in seven Chinese cities. Allergy. 2015;70:533–539. doi:10.1111/all.12577
3. Wang X, Zhang N, Bo M, et al. Diversity of TH cytokine profiles in patients with chronic rhinosinusitis: a multicenter study in Europe, Asia, and Oceania. J Allergy Clin Immunol. 2016;138:1344–1353. doi:10.1016/j.jaci.2016.05.041
4. Workman AD, Kohanski MA, Cohen NA. Biomarkers in chronic rhinosinusitis with nasal polyps. Immunol Allergy Clin North Am. 2018;38:679–692. doi:10.1016/j.iac.2018.06.006
5. Cao PP, Li H-B, Wang B-F, et al. Distinct immunopathologic characteristics of various types of chronic rhinosinusitis in adult Chinese. J Allergy Clin Immunol. 2009;124:
6. Akdis CA, Bachert C, Cingi C, et al. Endotypes and phenotypes of chronic rhinosinusitis: a PRACTALL document of the European Academy of Allergy and Clinical Immunology and the American Academy of Allergy, Asthma & Immunology. J Allergy Clin Immunol. 2013;131:1479–1490. doi:10.1016/j.jaci.2013.02.036
7. Meltzer EO, Hamilos DL, Hadley JA, et al. Rhinosinusitis: establishing definitions for clinical research and patient care. Otolaryngol Head Neck Surg. 2004;131:S1–S62. doi:10.1016/j.otohns.2004.09.067
8. Fokkens WJ, Lund VJ, Mullol J, et al. EPOS 2012: European position paper on rhinosinusitis and nasal polyps 2012. A summary for otorhinolaryngologists. Rhinology. 2012;50:1–12. doi:10.4193/Rhino12.000
9. Bachert C, Marple B, Schlosser RJ, et al. Adult chronic rhinosinusitis. Nat Rev Dis Primers. 2020;6:86. doi:10.1038/s41572-020-00218-1
10. Rosenfeld RM, Piccirillo JF, Chandrasekhar SS, et al. Clinical practice guideline (update): adult sinusitis. Otolaryngol Head Neck Surg. 2015;152:S1–S39. doi:10.1177/0194599815572097
11. Seshadri S, Lin DC, Rosati M, et al. Reduced expression of antimicrobial PLUNC proteins in nasal polyp tissues of patients with chronic rhinosinusitis. Allergy. 2012;67:920–928. doi:10.1111/j.1398-9995.2012.02848.x
12. Tantilipikorn P, Sompornrattanaphan M, Suwanwech T, Ngaotepprutaram P. Chronic rhinosinusitis and allergy: increased allergen sensitization versus real allergic rhinitis multimorbidity: a systematic review. Curr Allergy Asthma Rep. 2020;20:19. doi:10.1007/s11882-020-00913-9
13. Stevens WW, Schleimer RP, Kern RC. Chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol Pract. 2016;4:565–572. doi:10.1016/j.jaip.2016.04.012
14. Soyka MB, Wawrzyniak P, Eiwegger T, et al. Defective epithelial barrier in chronic rhinosinusitis: the regulation of tight junctions by IFN-gamma and IL-4. J Allergy Clin Immunol. 2012;130:1087–1096 e1010. doi:10.1016/j.jaci.2012.05.052
15. Tieu DD, Kern RC, Schleimer RP. Alterations in epithelial barrier function and host defense responses in chronic rhinosinusitis. J Allergy Clin Immunol. 2009;124:37–42. doi:10.1016/j.jaci.2009.04.045
16. Kao SS, Ramezanpour M, Bassiouni A, et al. Barrier disruptive effects of mucus isolated from chronic rhinosinusitis patients. Allergy. 2020;75:200–203. doi:10.1111/all.13964
17. Schleimer RP. Immunopathogenesis of chronic rhinosinusitis and nasal polyposis. Annu Rev Pathol. 2017;12:331–357. doi:10.1146/annurev-pathol-052016-100401
18. Zhang N, Van Crombruggen K, Gevaert E, Bachert C. Barrier function of the nasal mucosa in health and type-2 biased airway diseases. Allergy. 2016;71:295–307. doi:10.1111/all.12809
19. Schleimer RP, Kato A, Kern R, Kuperman D, Avila PC. Epithelium: at the interface of innate and adaptive immune responses. J Allergy Clin Immunol. 2007;120:1279–1284. doi:10.1016/j.jaci.2007.08.046
20. Van Bruaene N, Bachert C. Tissue remodeling in chronic rhinosinusitis. Curr Opin Allergy Clin Immunol. 2011;11:8–11. doi:10.1097/ACI.0b013e32834233ef
21. Barham HP, Osborn JL, Snidvongs K, et al. Remodeling changes of the upper airway with chronic rhinosinusitis. Int Forum Allergy Rhinol. 2015;5:565–572. doi:10.1002/alr.21546
22. Kern RC, Conley DB, Walsh W, et al. Perspectives on the etiology of chronic rhinosinusitis: an immune barrier hypothesis. Am J Rhinol. 2008;22:549–559. doi:10.2500/ajr.2008.22.3228
23. Meng J, Zhou P, Liu Y, et al. The development of nasal polyp disease involves early nasal mucosal inflammation and remodelling. PLoS One. 2013;8:e82373. doi:10.1371/journal.pone.0082373
24. Brozek JL, Bousquet J, Agache I, et al. Allergic rhinitis and its impact on asthma (ARIA) guidelines-2016 revision. J Allergy Clin Immunol. 2017;140:950–958. doi:10.1016/j.jaci.2017.03.050
25. Jang YJ, Kim HG, Koo TW, Chung PS. Localization of ZO-1 and E-cadherin in the nasal polyp epithelium. Eur Arch Otorhinolaryngol. 2002;259:465–469. doi:10.1007/s00405-002-0500-z
26. Rogers GA, Beste KD, Parkos CA, et al. Epithelial tight junction alterations in nasal polyposis. Int Forum Allergy Rhinol. 2011;1:50–54. doi:10.1002/alr.20014
27. Barmeyer C, Schulzke JD, Fromm M. Claudin-related intestinal diseases. Semin Cell Dev Biol. 2015;42:30–38. doi:10.1016/j.semcdb.2015.05.006
28. Shahana S, Jaunmuktane Z, Asplund MS, Roomans GM. Ultrastructural investigation of epithelial damage in asthmatic and non-asthmatic nasal polyps. Respir Med. 2006;100:2018–2028. doi:10.1016/j.rmed.2006.02.012
29. Loxham M, Davies DE. Phenotypic and genetic aspects of epithelial barrier function in asthmatic patients. J Allergy Clin Immunol. 2017;139:1736–1751. doi:10.1016/j.jaci.2017.04.005
30. Georas SN, Rezaee F. Epithelial barrier function: at the front line of asthma immunology and allergic airway inflammation. J Allergy Clin Immunol. 2014;134:509–520. doi:10.1016/j.jaci.2014.05.049
31. Yu XM, Li CW, Chao SS, et al. Reduced growth and proliferation dynamics of nasal epithelial stem/progenitor cells in nasal polyps in vitro. Sci Rep. 2014;4:4619. doi:10.1038/srep04619
32. Yu XM, Li CW, Li YY, et al. Down-regulation of EMP1 is associated with epithelial hyperplasia and metaplasia in nasal polyps. Histopathology. 2013;63:686–695. doi:10.1111/his.12211
33. Wang W, Gao Z, Wang H, et al. Transcriptome analysis reveals distinct gene expression profiles in eosinophilic and noneosinophilic chronic rhinosinusitis with nasal polyps. Sci Rep. 2016;6:26604. doi:10.1038/srep26604
34. Ma Y, Zheng C, Shi L. The kinase LRRK2 is differently expressed in chronic rhinosinusitis with and without nasal polyps. Clin Transl Allergy. 2018;8:8. doi:10.1186/s13601-018-0194-y
35. Nian JB, Zeng M, Zheng J, et al. Epithelial cells expressed IL-33 to promote degranulation of mast cells through inhibition on ST2/PI3K/mTOR-mediated autophagy in allergic rhinitis. Cell Cycle. 2020;19:1132–1142. doi:10.1080/15384101.2020.1749402
36. Wlodarska M, Thaiss C, Nowarski R, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–1059. doi:10.1016/j.cell.2014.01.026
37. Buque A, Bloy N, Perez-Lanzón M, et al. Immunoprophylactic and immunotherapeutic control of hormone receptor-positive breast cancer. Nat Commun. 2020;11:3819. doi:10.1038/s41467-020-17644-0
38. Lai X, Li X, Chang L, et al. IL-19 up-regulates mucin 5AC production in patients with chronic rhinosinusitis via STAT3 pathway. Front Immunol. 2019;10:1682. doi:10.3389/fimmu.2019.01682
39. Cheng J, Chen J, Zhao Y, et al. MicroRNA-761 suppresses remodeling of nasal mucosa and epithelial-mesenchymal transition in mice with chronic rhinosinusitis through LCN2. Stem Cell Res Ther. 2020;11:151. doi:10.1186/s13287-020-01598-7
40. Shi S, Han Y, Wang D, et al. PD-L1 and PD-1 expressed in trigeminal ganglia may inhibit pain in an acute migraine model. Cephalalgia. 2020;40:288–298. doi:10.1177/0333102419883374
41. Parra ER, Villalobos P, Behrens C, et al. Effect of neoadjuvant chemotherapy on the immune microenvironment in non-small cell lung carcinomas as determined by multiplex immunofluorescence and image analysis approaches. J Immunother Cancer. 2018;6:48. doi:10.1186/s40425-018-0368-0
42. Van Bruaene N, Pérez-Novo CA, Basinski TM, et al. T-cell regulation in chronic paranasal sinus disease. J Allergy Clin Immunol. 2008;121:
43. Bachert C, Zhang N, Holtappels G, et al. Presence of IL-5 protein and IgE antibodies to staphylococcal enterotoxins in nasal polyps is associated with comorbid asthma. J Allergy Clin Immunol. 2010;126:
44. Tan BK, Klingler AI, Poposki JA, et al. Heterogeneous inflammatory patterns in chronic rhinosinusitis without nasal polyps in Chicago, Illinois. J Allergy Clin Immunol. 2017;139:699–703 e697. doi:10.1016/j.jaci.2016.06.063
45. Thanasumpun T, Batra PS. Endoscopically-derived bacterial cultures in chronic rhinosinusitis: a systematic review. Am J Otolaryngol. 2015;36:686–691. doi:10.1016/j.amjoto.2015.04.010
46. Zhang Z, Adappa ND, Doghramji LJ, et al. Different clinical factors associated with Staphylococcus aureus and Pseudomonas aeruginosa in chronic rhinosinusitis. Int Forum Allergy Rhinol. 2015;5:724–733. doi:10.1002/alr.21532
47. Corriveau MN, Zhang N, Holtappels G, Van Roy N, Bachert C. Detection of Staphylococcus aureus in nasal tissue with peptide nucleic acid-fluorescence in situ hybridization. Am J Rhinol Allergy. 2009;23:461–465. doi:10.2500/ajra.2009.23.3367
48. Tan NC, Foreman A, Jardeleza C, et al. Intracellular Staphylococcus aureus: the Trojan horse of recalcitrant chronic rhinosinusitis? Int Forum Allergy Rhinol. 2013;3:261–266. doi:10.1002/alr.21154
49. Kim RJ, Yin T, Chen C-J-J, et al. The interaction between bacteria and mucosal immunity in chronic rhinosinusitis: a prospective cross-sectional analysis. Am J Rhinol Allergy. 2013;27:e183–e189. doi:10.2500/ajra.2013.27.3974
50. Valera FCP, Ruffin M, Adam D, et al. Staphylococcus aureus impairs sinonasal epithelial repair: effects in patients with chronic rhinosinusitis with nasal polyps and control subjects. J Allergy Clin Immunol. 2019;143:591–603 e593. doi:10.1016/j.jaci.2018.05.035
51. Tan NC, Cooksley CM, Roscioli E, et al. Small-colony variants and phenotype switching of intracellular Staphylococcus aureus in chronic rhinosinusitis. Allergy. 2014;69:1364–1371. doi:10.1111/all.12457
52. Kohanski MA, Lane AP. Sinonasal epithelial cell response to Staphylococcus aureus burden in chronic rhinosinusitis. JAMA Otolaryngol Head Neck Surg. 2015;141:341–349. doi:10.1001/jamaoto.2014.3550
53. Gevaert E, Zhang N, Krysko O, et al. Extracellular eosinophilic traps in association with Staphylococcus aureus at the site of epithelial barrier defects in patients with severe airway inflammation. J Allergy Clin Immunol. 2017;139:1849–1860 e1846. doi:10.1016/j.jaci.2017.01.019
54. Teufelberger AR, Nordengrün M, Braun H, et al. The IL-33/ST2 axis is crucial in type 2 airway responses induced by Staphylococcus aureus-derived serine protease-like protein D. J Allergy Clin Immunol. 2018;141:549–559 e547. doi:10.1016/j.jaci.2017.05.004
55. Malik Z, Roscioli E, Murphy J, et al. Staphylococcus aureus impairs the airway epithelial barrier in vitro. Int Forum Allergy Rhinol. 2015;5(6):551–556. doi:10.1002/alr.21517
56. Kim CS, Jeon S-Y, Min Y-G, et al. Effects of beta-toxin of Staphylococcus aureus on ciliary activity of nasal epithelial cells. Laryngoscope. 2000;110:2085–2088. doi:10.1097/00005537-200012000-00021
57. Eiffler I, Behnke J, Ziesemer S, Muller C, Hildebrandt JP. Staphylococcus aureus alpha-toxin-mediated cation entry depolarizes membrane potential and activates p38 MAP kinase in airway epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2016;311:L676–L685. doi:10.1152/ajplung.00090.2016
58. Hermann I, Räth S, Ziesemer S, et al. Staphylococcus aureus hemolysin A disrupts cell–matrix adhesions in human airway epithelial cells. Am J Respir Cell Mol Biol. 2015;52:14–24. doi:10.1165/rcmb.2014-0082OC
59. LeMessurier KS, Tiwary M, Morin NP, Samarasinghe AE. Respiratory barrier as a safeguard and regulator of defense against influenza A virus and Streptococcus pneumoniae. Front Immunol. 2020;11:3. doi:10.3389/fimmu.2020.00003
60. Burgel PR, Nadel JA. Roles of epidermal growth factor receptor activation in epithelial cell repair and mucin production in airway epithelium. Thorax. 2004;59:992–996. doi:10.1136/thx.2003.018879
61. Jacob M, Bin Khalaf D, Alhissi S, et al. Quantitative profiling of cytokines and chemokines in DOCK8-deficient and atopic dermatitis patients. Allergy. 2019;74:370–379. doi:10.1111/all.13610
62. Pastore S, Mascia F, Mariani V, Girolomoni G. The epidermal growth factor receptor system in skin repair and inflammation. J Invest Dermatol. 2008;128:1365–1374. doi:10.1038/sj.jid.5701184
63. Sinha A, Nightingale J, West KP, Berlanga-Acosta J, Playford RJ. Epidermal growth factor enemas with oral mesalamine for mild-to-moderate left-sided ulcerative colitis or proctitis. N Engl J Med. 2003;349:350–357. doi:10.1056/NEJMoa013136
64. Brown GL, Nanney LB, Griffen J, et al. Enhancement of wound healing by topical treatment with epidermal growth factor. N Engl J Med. 1989;321:76–79. doi:10.1056/NEJM198907133210203
65. Duan C, Li CW, Zhao L, et al. Differential expression patterns of EGF, EGFR, and ERBB4 in nasal polyp epithelium. PLoS One. 2016;11:e0156949. doi:10.1371/journal.pone.0156949
66. Ding GQ, Zheng CQ, Bagga SS. Up-regulation of the mucosal epidermal growth factor receptor gene in chronic rhinosinusitis and nasal polyposis. Arch Otolaryngol Head Neck Surg. 2007;133:1097–1103. doi:10.1001/archotol.133.11.1097
67. Marinas AE, Ciurea P, Margaritescu C, Cotoi OS. Expression of epidermal growth factor (EGF) and its receptors (EGFR1 and EGFR2) in chronic bronchitis. Rom J Morphol Embryol. 2012;53:957–966.
68. Sheng W, Shi X, Lin Y, et al. Musashi2 promotes EGF-induced EMT in pancreatic cancer via ZEB1-ERK/MAPK signaling. J Exp Clin Cancer Res. 2020;39:16. doi:10.1186/s13046-020-1521-4
69. Okuyama M, Fujiwara Y, Tanigawa T, et al. Roles of ZO-1 and epidermal growth factor in esophageal epithelial defense against acid. Digestion. 2007;75:135–141. doi:10.1159/000106454
70. Kugler MC, Joyner AL, Loomis CA, Munger JS. Sonic hedgehog signaling in the lung. From development to disease. Am J Respir Cell Mol Biol. 2015;52:1–13. doi:10.1165/rcmb.2014-0132TR
71. Konigshoff M, Balsara N, Pfaff E-M, et al. Functional Wnt signaling is increased in idiopathic pulmonary fibrosis. PLoS One. 2008;3:e2142. doi:10.1371/journal.pone.0002142
72. Choy DF, Modrek B, Abbas AR, et al. Gene expression patterns of Th2 inflammation and intercellular communication in asthmatic airways. J Immunol. 2011;186:1861–1869. doi:10.4049/jimmunol.1002568
73. Chilosi M, Poletti V, Zamò A, et al. Aberrant Wnt/beta-catenin pathway activation in idiopathic pulmonary fibrosis. Am J Pathol. 2003;162:1495–1502. doi:10.1016/s0002-9440(10)64282-4
74. Aumiller V, Balsara N, Wilhelm J, Gunther A, Konigshoff M. WNT/beta-catenin signaling induces IL-1beta expression by alveolar epithelial cells in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2013;49:96–104. doi:10.1165/rcmb.2012-0524OC
75. Konigshoff M, Kramer M, Balsara N, et al. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J Clin Invest. 2009;119:772–787. doi:10.1172/JCI33950
76. Konigshoff M, Eickelberg O. WNT signaling in lung disease: a failure or a regeneration signal? Am J Respir Cell Mol Biol. 2010;42:21–31. doi:10.1165/rcmb.2008-0485TR
77. Halleskog C, Mulder J, Dahlström J, et al. WNT signaling in activated microglia is proinflammatory. Glia. 2011;59:119–131. doi:10.1002/glia.21081
78. Sen M, Lauterbach K, El-Gabalawy H, et al. Expression and function of wingless and frizzled homologs in rheumatoid arthritis. Proc Natl Acad Sci U S A. 2000;97:2791–2796. doi:10.1073/pnas.050574297
79. Blumenthal A, Ehlers S, Lauber J, et al. The Wingless homolog WNT5A and its receptor Frizzled-5 regulate inflammatory responses of human mononuclear cells induced by microbial stimulation. Blood. 2006;108:965–973. doi:10.1182/blood-2005-12-5046
80. Boscke R, Vladar EK, Könnecke M, et al. Wnt signaling in chronic rhinosinusitis with nasal polyps. Am J Respir Cell Mol Biol. 2017;56:575–584. doi:10.1165/rcmb.2016-0024OC
81. Whyte JL, Smith AA, Helms JA. Wnt signaling and injury repair. Cold Spring Harb Perspect Biol. 2012;4:a008078. doi:10.1101/cshperspect.a008078
82. Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18:246–254. doi:10.1038/ncb3312
83. Hsu YC, Li L, Fuchs E. Emerging interactions between skin stem cells and their niches. Nat Med. 2014;20:847–856. doi:10.1038/nm.3643
84. Lindemans CA, Calafiore M, Mertelsmann AM, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528:560–564. doi:10.1038/nature16460
85. Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi:10.1038/nature06196
86. Sato T, van Es JH, Snippert HJ, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi:10.1038/nature09637
87. Bielefeld KA, Amini-Nik S, Alman BA. Cutaneous wound healing: recruiting developmental pathways for regeneration. Cell Mol Life Sci. 2013;70:2059–2081. doi:10.1007/s00018-012-1152-9
88. Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453:314–321. doi:10.1038/nature07039
89. Knight C, James S, Kuntin D, et al. Epidermal growth factor can signal via beta-catenin to control proliferation of mesenchymal stem cells independently of canonical Wnt signalling. Cell Signal. 2019;53:256–268. doi:10.1016/j.cellsig.2018.09.021
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.