Back to Journals » International Journal of Nanomedicine » Volume 10 » Issue 1
Efficient gene delivery to human umbilical cord mesenchymal stem cells by cationized Porphyra yezoensis polysaccharide nanoparticles
Authors Yu Q, Cao J, Chen B , Deng W, Cao X , Chen J, Wang Y, Wang S, Yu J, Xu X, Gao X
Received 25 July 2015
Accepted for publication 3 October 2015
Published 18 November 2015 Volume 2015:10(1) Pages 7097—7107
DOI https://doi.org/10.2147/IJN.S93122
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Lei Yang
Qingtong Yu,1,2 Jin Cao,2 Baoding Chen,3 Wenwen Deng,2 Xia Cao,2 Jingjing Chen,2 Yan Wang,2 Shicheng Wang,2 Jiangnan Yu,2 Ximing Xu,2 Xiangdong Gao1
1School of Life Science and Technology, China Pharmaceutical University, Nanjing, 2Department of Pharmaceutics, School of Pharmacy and Center for Drug/Gene Delivery and Tissue Engineering, Jiangsu University, 3Department of Medical Ultrasound, Affiliated Hospital of Jiangsu University, Zhenjiang, People’s Republic of China
Abstract: This study centered on an innovative application of Porphyra yezoensis polysaccharide (PPS) with cationic modification as a safe and efficient nonviral gene vector to deliver a plasmid encoding human Wnt3a (pWnt3a) into human umbilical cord mesenchymal stem cells (HUMSCs). After modification with branched low-molecular-weight (1,200 Da) polyethylenimine, the cationized PPS (CPPS) was combined with pWnt3a to form spherical nanoscale particles (CPPS-pWnt3a nanoparticles). Particle size and distribution indicated that the CPPS-pWnt3a nanoparticles at a CPPS:pWnt3a weight ratio of 40:1 might be a potential candidate for DNA plasmid transfection. A cytotoxicity assay demonstrated that the nanoparticles prepared at a CPPS:pWnt3a weight ratio of 40:1 were nontoxic to HUMSCs compared to those of Lipofectamine 2000 and polyethylenimine (25 kDa). These nanoparticles were further transfected to HUMSCs. Western blotting demonstrated that the nanoparticles (CPPS:pWnt3a weight ratio 40:1) had the greatest transfection efficiency in HUMSCs, which was significantly higher than that of Lipofectamine 2000; however, when the CPPS:pWnt3a weight ratio was increased to 80:1, the nanoparticle-treated group showed no obvious improvement in translation efficiency over Lipofectamine 2000. Therefore, CPPS, a novel cationic polysaccharide derived from P. yezoensis, could be developed into a safe, efficient, nonviral gene vector in a gene-delivery system.
Keywords: Porphyra yezoensis, nanoparticle, nonviral vector, human umbilical cord mesenchymal stem cells, Wnt3a
Introduction
Porphyra yezoensis, widely distributed in eastern Asia and on the west coast of the USA, has been praised as an “affinal drug and diet” for its various biological activities. Edible P. yezoensis, commonly referred to as nori,1 has become a promising source of nutrients, owing to its richness in polysaccharides, which have antioxidative,2–5 anti-inflammatory,6,7 hyperglycemic,8 and immunostimulatory effects.9 More recently, several studies have revealed that sulfated polysaccharides from P. yezoensis can inhibit tumor growth both in vitro and in vivo.10,11 However, despite its biological and pharmacological functions, little attention has been paid to P. yezoensis polysaccharide (PPS) as a functional material for gene transfection.
With the tremendous progress in biotechnology, gene therapy has been proposed as a strategy for the treatment of some diseases, such as incurable cancers. Successful delivery of a specific functional gene in a simple manner is vital in gene therapy. Given the chance of degradation of naked therapeutic genes by nucleases and the low efficiency of the cellular uptake, the development of efficient and safe gene vectors is an urgent need.12,13 Recombinant viruses, such as lentivirus,14–18 retrovirus,19–22 and adenovirus,23–25 have been considered ideal gene carriers because of their considerable cellular uptake. Even though their high transfection efficiency might make clinical applications of recombinant viruses possible in gene therapy, obstacles, such as abnormal gene expression26–28 and immune responses,29–31 are still unresolved for the use of viral vectors.
Nonviral vectors, which have been regarded as alternative gene carriers for functional gene delivery, have received a great deal of attention, due to their advanced properties, such as low immunogenicity, safety, and specificity, as well as long-term gene expression. Cationic lipids, such as Lipofectamine32,33 and 1,2-dioleoyl-3-trimethylammonium,34,35 are popular nonviral vectors that are widely used for target-gene delivery because of their high in vitro transfection efficiency. In addition, cationic polymers that contain amino groups in their side chains or backbones have become another promising nonviral gene carrier, not only because of their ease of production, low risk of side effects, and non-immunogenicity36 but also for their compact combination with DNA,37 which is an issue of crucial importance during gene transfection. However, these cationic polyplexes also have inevitable drawbacks that restrict their clinical application. For example, cytotoxicity and macrophage activation caused by cationic liposomes may induce cell shrinkage and vacuolization of the cytoplasm.38 To overcome these difficulties, polysaccharides, a natural product and an ideal material, have been modified and reformed to produce gene vectors. Ethylenediamine-modified polysaccharide from mulberry leaves39 and Lycium babarum40 has high efficiency for the transfer of a DNA plasmid encoding TGFβ1 into rat bone marrow mesenchymal stem cells. Moreover, Angelica sinensis polysaccharide modified with polyethylenimine41 and Pleurotus eryngii polysaccharide cationized by spermine42 could become potential candidate nonviral vectors for gene transfection, because of their low cytotoxicity and excellent transfection efficiency compared to Lipofectamine 2000.
The objective of this study was to establish a novel gene vector, cationized PPS (CPPS) nanoparticles, for the nonviral gene delivery of a plasmid DNA encoding Wnt3a into human umbilical cord mesenchymal stem cells (HUMSCs) in vitro. Specifically, we optimized the extraction process for PPS and successfully developed a CPPS via chemical modification with low-molecular-weight polyethylenimine (PEI; 600 Da). Moreover, HUMSCs, which normally would otherwise be discarded after birth, have emerged as a desirable source of multipotent progenitor cells that can differentiate into several lineage-specific cell types to form bone, fat, tendon, and muscle tissue.39 Meanwhile, HUMSCs have been ultimately converted into induced pluripotent stem cells via the codelivery of Yamanaka factors (c-Myc, Klf4, Sox2, and Oct4) using calcium phosphate nanoparticles,43 which illustrated the remarkable inducibility of HUMSCs based on a nonviral gene-delivery system. In addition, the overexpression of Wnt3a44 can direct the differentiation of neural stem cells into neurons both in vitro and in vivo by activating Wnt signaling, which is also a requirement for the maintenance, proliferation, and survival of neural progenitors when cultured under neurosphere conditions.45 Previous studies have demonstrated that the overexpression of Wnt3a in highly self-renewable cells46,47 might contribute to the formation and proliferation of neural cells, which could be a new and safe method for the recovery of spinal cord injury.48,49 The novelty of this study is that we established a CPPS for the first time as a nonviral vector for the delivery of plasmid Wnt3a (pWnt3a) into HUMSCs, which might facilitate the effective application of P. yezoensis to the area of gene therapy.
Materials and methods
Materials
PPS was obtained in our laboratory.8 Branched PEI (molecular weight 600 Da) was purchased from Sigma-Aldrich (St Louis, MO, USA). Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium was purchased from Thermo Fisher Scientific (Waltham, MA, USA), as were penicillin–streptomycin, trypsin, fetal bovine serum (FBS), Lipofectamine 2000, and MTT. Antibodies, including mouse polyclonal anti-GAPDH, were purchased from Santa Cruz Biotechnology Inc (Dallas, TX, USA). Rabbit polyclonal anti-Wnt3a primary antibody was obtained from Abcam (Cambridge, UK). Horseradish peroxidase-conjugated secondary antibodies were obtained from Bioworld Technology, Inc., (Wuhan, People’s Republic of China [PRC]). All other chemicals and reagents were of an analytical grade or better, and were used without further purification. All of the solutions were prepared with highly purified water. The experimental protocol was approved by the University Ethics Committee for the Use of Experimental Animals, and it conformed to the Guide for Care and Use of Laboratory Animals. The experiments involving human cell lines were approved by the Institutional Review Board of Jiangsu University (Zhenjiang, China).
Preparation of CPPS
Extraction and purification of PPS
The purification of PPS in our laboratory8 was conducted according to a procedure illustrated in Figure 1. In brief, dried P. yezoensis powder (100 g) was homogenized and dispersed in 1 L of 95% ethanol and refluxed in a hot water bath at 70°C for 1 hour, followed by filtration to obtain the residue. Then, the residue was decentralized in double-distilled water (DDW; solid:liquid ratio 1:30, pH 11) at 90°C for 3 hours, followed by filtration. This process was repeated once. The filtrates from each run were combined and concentrated under reduced pressure at 80°C and precipitated with 95% (v/v) ethanol until the final ethanol concentration reached 80% (v/v). The resulting precipitate was dissolved into 150 mL DDW and reprecipitated with absolute ethanol. Finally, the ultimate precipitate was lyophilized to generate a crude extract, which was dissolved in DDW and applied to a diethylaminoethanol cellulose (DE-52; Shanghai YuanYe Biotechnology Co Ltd, Shanghai, PRC) column (diameter 2.6 cm × length 40 cm) and a Sephadex G-100 gel resin (Shanghai Richu Bioscience Co Ltd, Shanghai, PRC) column (diameter 2.6 cm × length 40 cm) in sequence. The fractions that contained polysaccharides were detected with a phenol–sulfuric acid assay. A single elution peak was chosen, collected, concentrated, dialyzed, and lyophilized to obtain purified PPS.
  | Figure 1 Flowchart for Porphyra yezoensis polysaccharide extraction and purification. |
Preparation of CPPS
As shown in Figure 2, 112 mg of PPS was dissolved in 10 mL of phosphate-buffered saline (PBS; pH 7.0) in a round-bottom flask that was degassed with N2. N, N′-carbonyldiimidazole (80 mg) was dissolved in 4 mL of methylene chloride. After the addition of 0.15 mL of triethylamine (Et2N) to the reaction, the methylene chloride–carbonyldiimidazole solution was added to the reaction slowly with gentle magnetic stirring. After 60 minutes of reaction time, a PEI (600 Da) solution (1,200 mg PEI dissolved in 15 mL PBS, pH 7.0) was slowly added to the reaction mixture and magnetically stirred in a dark room for 24 hours. Finally, the mixture was dialyzed (molecular weight cutoff 3,000 Da; Genia Biotech, Beijing, PRC) against DDW for 2 days and then freeze-dried to yield CPPS.
  | Figure 2 Flowchart for cationized Porphyra yezoensis polysaccharide preparation. |
Cell culture
HUMSCs, generously provided by the Beike Jiangsu Stem Cell Bank (Taizhou, Jiangsu, PRC), were cultured in DMEM/F12 supplemented with 10% FBS, 0.1 mM nonessential amino acids (NEAAs; Sigma-Aldrich), FGFβ (PeproTech, Rocky Hill, NJ, USA), and 0.1 mM β-mercaptoethanol (β-ME; Toyoda Bio, Beijing, PRC). The cells were passaged with 0.25% trypsin and 0.1% ethylenediaminetetraacetic acid (EDTA) and incubated at 37°C in 5% humidified CO2.
Preparation of CPPS-pWnt3a complexes
CPPS-pWnt3a complexes were prepared by coacervation,50 a technique based on the electrostatic interaction of two oppositely charged compounds. Briefly, 20 mg of PPS-PEI was fully dissolved in 1 mL of sterile water to obtain a PPS-PEI stock solution (20 mg/mL). The working pWnt3a solution (20 μg/mL) was prepared with sterile water. Aliquots (100 μL) of CPPS solution and pWnt3a were heated separately at 55°C for 30 minutes. Equal volumes of these two solutions were immediately mixed together and vortexed for 30 seconds to obtain CPPS-Wnt3a nanoparticles and then incubated at room temperature for 30 minutes.
Electrophoresis of PPS-Wnt3a nanoparticles
The plasmid DNA retardation effect of the CPPS-Wnt3a nanoparticles was analyzed using 1% agarose gel electrophoresis. Different PPS:Wnt3a weight ratios (5:1, 10:1, 20:1, 40:1, 80:1, 100:1, and 150:1) of CPPS-Wnt3a nanoparticles were prepared. The PPS-Wnt3a nanoparticle solution (5 μL) was added to 1 μL of loading buffer (0.1% sodium dodecyl sulfate, 5% glycerol, and 0.005% bromophenol blue) and applied to a 1% agarose gel in a Tris/borate/EDTA (TBE) buffer solution (pH 8.0) containing 0.1 mg·mL−1 ethidium bromide. Meanwhile, 5 μL of a naked pWnt3a solution that was diluted with sterile water was used as a negative control. Electrophoresis of the nanoparticles was performed in TBE solution at 80 V for 90 minutes. The gel was imaged with an ultraviolet transilluminator (Gel Doc 2000; Bio-Rad Laboratories Inc, Hercules, CA, USA).
Cytotoxicity assay
The in vitro cytotoxicity of the CPPS-Wnt3a complex was measured by the MTT assay. Briefly, the HUMSCs were seeded in 96-well plates at a density of 30,000 cells per well and incubated at 37°C. The medium was subsequently removed and replaced with serum-free medium until cell confluence reached 80%. After 12 hours of serum-free incubation, CPPS-Wnt3a nanoparticles (weight ratios of 40:1, 80:1, 100:1, 150:1, 200:1, 250:1, and 300:1) were transferred, and the Lipofectamine 2000-Wnt3a and PEI-Wnt3a groups (multiple dilutions of PEI: 50-fold, 100-fold, 200-fold, 400-fold, and 800-fold) were used as positive controls according to the manufacturer’s protocol under the same conditions. The viability percentage was relative to untreated cells. The cell viability (%) was calculated by the formula cell viability (%) = Abssample/Abscontrol ×100.
Zeta potential
The zeta potential of the nanoparticle suspensions was measured with a Nano ZS90 Zetasizer (Malvern Instruments, Malvern, UK). The zeta potential of the free plasmid and the original CPPS were determined under the same conditions.
Determination of nanoparticle-size distribution
The nanoparticle-size distribution was determined by a dynamic light-scattering technique, performed at 25°C with the Nano ZS90 Zetasizer. The measured scattering intensity was then analyzed by the software provided by Malvern.
Transmission electron microscopy
Transmission electron microscopy (TEM; Tecnai 12; Philips, Amsterdam, the Netherlands) was used to investigate the shape and size of the PPS-PEI/Wnt3a. Briefly, 1 μL of nanoparticle suspension was applied to a copper screen and air-dried. The air-dried samples were then observed directly under TEM.
Gene transfection
GFP transfection
HUMSCs were seeded in 24-well plates at a density of 100,000 cells/well in 0.5 mL of DMEM/F12 containing 10% FBS without antibiotics and cultured for 24 hours to reach a confluence of 80% before gene transfection. The medium was subsequently replaced with 0.5 mL of serum-free DMEM/F12 at 12 hours before transfection. After removal of the medium, 0.5 mL of the nanoparticle suspension (pGFP 800 ng/well) prepared in three different CPPS:pGFP weight ratios (40:1, 80:1, and 100:1) was added to each well. The Lipofectamine 2000–pGFP complex was used as a positive control according to the manufacturer’s instructions. The medium was replaced with fresh growth medium (DMEM/F12 containing 10% FBS, 0.1 mM NEAA, 0.1 mM β-ME, and FGF-β) 5 hours after transfection. All cells were cultured at 37°C for 72 hours after transfection.
pWnt3a transfection
The HUMSCs were seeded in six-well plates at a density of 400,000 cells per well in 2 mL DMEM/F12 containing 10% FBS without antibiotics and cultured for 24–48 hours to reach a confluence of 80% before gene transfection. The medium was subsequently replaced with 2 mL of serum-free DMEM/F12 at 12 hours before transfection. After removal of the medium, 2 mL of the nanoparticle suspension (pWnt3a 4 μg/well) diluted with serum-free medium was added to each well. The Lipofectamine 2000–Wnt3a complex was used as a positive control according to the manufacturer’s instructions. Wnt3a free plasmid in a serum-free nutrient solution was used as a negative control. Four hours after transfection, the medium was replaced with fresh growth medium (DMEM/F12 containing 10% FBS, 0.1 mM NEAA, 0.1 mM β-ME, and FGF-β). All cells were cultured at 37°C for 72 hours after transfection.
Western blotting
The transfected HUMSCs were lysed in radioimmunoprecipitation assay lysis buffer with a protease-inhibitor cocktail (Santa Cruz Biotechnology) in an ice bath for 30 minutes, then washed with PBS and sonicated for 1 minute. The cell lysates were harvested from the supernatant after centrifugation at 12,000 ×g for 5 minutes. The protein extracts were mixed with an equal volume of loading buffer and then heat-denatured at 100°C for 5 minutes. An equal amount of protein from each sample was loaded onto 12% sodium dodecyl sulfate polyacrylamide gels, separated electrophoretically, and transferred to a polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA) by electrophoresis. The polyvinylidene difluoride membrane was incubated with antibodies against human Wnt3a and GADPH (standard control), followed by horseradish peroxidase-conjugated secondary antibodies. Protein bands were visualized using a Pierce Eclipse Plus substrate (Thermo Fisher Scientific) and scanned with a ChemiScope 3200 mini (Shanghai Clinx Science Instruments Co Ltd, Shanghai, PRC).
Statistical analyses
All data are reported as means ± standard deviation. Two-way analysis of variance was used to analyze all data in this study with GraphPad Prism version 5.01 for Windows (GraphPad Software Inc, La Jolla, CA, USA). P<0.05 was considered statistically significant.
Results and discussion
CPPS characteristics
Purified PPS was made in our laboratory. Fourier-transform infrared (FTIR) analysis was carried out to characterize the CPPS through at least three repeated operations. A qualitative functional group analysis by FTIR (KBr) of PEI-modified PPS yielded peaks at 3,410, 1,650, 1,250, and 1,070 cm−1. In Figure 3, compared with the spectrum of PPS, the strong absorption peak at 3,410 cm−1 was assigned to –NH stretching. The absorption peak at 2,935 cm−1 of PEI-PPS, which was stronger than that in PPS, was the absorption peak of methylene. The absorption peak at 1,650 cm−1 indicated the in-plane and out-of-plane bending vibration of β-amides. In addition, the absorption peaks at 1,250 and 1,070 cm−1 were significantly stronger than those in PPS, which we attributed to C–N stretching. All of the absorption peaks demonstrated that PPS was successfully cationized by PEI through the connection between –CO–NH– and R2NH of PEI.
  | Figure 3 Fourier-transform infrared spectra of Porphyra yezoensis polysaccharide (PPS) and polyethylenimine (PEI)-PPS. |
Agarose gel electrophoresis assay
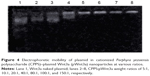
The combination of CPPS and plasmid DNA was evaluated at various CPPS:pWnt3a weight ratios (5:1, 10:1, 20:1, 40:1, 80:1, 100:1, and 150:1) with an agarose gel retardation assay, which was conducted in triplicate (Figure 4). An increase in the CPPS:pWnt3a weight ratio retarded the DNA migration. No retardation was detected for the complexes at CPPS:pWnt3a weight ratios of 5:1 or 10:1 (lanes 2 and 3 in Figure 4). Partial migration of the plasmids was observed with the complex that was prepared at a CPPS:pWnt3a weight ratio of 20:1 (lane 4 in Figure 4). In contrast, no plasmid migration occurred when the CPPS:pWnt3a weight ratio reached 40:1, which illustrated that the plasmids were retarded completely. This result indicated that the firmness of the combination of CPPS and DNA plasmid was highly associated with the content of the CPPS in the CPPS-pWnt3a nanoparticles. A higher CPPS:pWnt3a weight ratio resulted in a superior combination of CPPS and pWnt3a.
Cytotoxicity assay of CPPS-pWnt3a nanoparticles
Low cytotoxicity is crucial for evaluating the safety of a gene carrier during in vitro transfection. Based on the results of the gel retardation assay, the CPPS:pWnt3a weight ratio for testing was set from 40:1 to 300:1. In Figure 5, the MTT assay revealed that a higher CPPS:pWnt3a ratio led to increased cytotoxicity. No obvious differences in cytotoxicity occurred at CPPS:pWnt3a weight ratios of 40:1 and 80:1 compared to the free pWnt3a group, which was used as a negative control. However, in the Wnt3a group, cell viability was slightly higher than 100%, which may have been due to experimental errors. Meanwhile, as for the MCPPS/MpWnt3a (40:1) group, the increased cell viability might have been due to the excellent cell compatibility of the cationic polysaccharide, which may favor cell proliferation to a certain extent. In addition, the percentage of viable cells at a CPPS:pWnt3a ratio of 100:1 and 150:1, higher ratios than for the positive control Lipofectamine 2000, was distinctly lower than that in CPPS:pWnt3a weight ratios of 40:1 and 80:1. In addition, the PEI group displayed a continuous increase in cell viability with a reduction in PEI concentration, which indicated that PEI cannot be used alone as a gene vector for gene delivery because of its high cytotoxicity.
A possible reason for this result is that PEI, which is frequently utilized for cationic modification, might induce the overexpression of caveolin 1, which could partially determine the transfection efficiency and the cytotoxicity of PEI.51 Similarly, with an increase in the CPPS:pWnt3a weight ratio, the concentration of PEI also increased during the gene transfection, which led to a gradual increase in cytotoxicity.
Zeta-potential measurement
Zeta-potential measurement is a common method to test the ability of a gene vector to combine with the negatively charged plasmid. In addition, positively charged nanoparticles may easily come into contact with negatively charged cytomembrane. According to the results of the gel retardation assay and the cytotoxicity test, the zeta potential of the nanoparticles, which were prepared at three different CPPS:pWnt3a weight ratios (40:1, 80:1, and 100:1), was measured with the Nano ZS90 Zetasizer. The zeta potential of the original CPPS and the free pWnt3a was also investigated under the same conditions. Figure 6 shows that the zeta potential of the original CPPS and Wnt3a plasmids was +16.9±0.6 and −38.7±1.5 mV, respectively. After cationic modification, the zeta potential of the nanoparticles, which were prepared at three different CPPS:pWnt3a weight ratios (40:1, 80:1, and 100:1), changed from the negative value of −38.7±1.5 mV to a positive value, and increased to 8.7±0.1, 10.8±0.3, and 13.3±1.0 mV, respectively. An increase in the amount of CPPS leading to an increased positive charge density provided solid evidence for the successful cationization of PPS.
Morphology and particle size of CPPS-pWnt3a nanoparticles
The size of the nanoparticles with CPPS:pWnt3a weight ratios of 40:1, 80:1, and 100:1 was investigated with the ZS90. The particle diameters, as shown in Figure 7, increased from 167.5±6.2 to 341.7±7.2 nm when the CPPS:pWnt3a weight ratio was increased from 40:1 to 100:1. The complex that was prepared at a CPPS:pWnt3a ratio of 40:1 obviously had the smallest particles. This could be attributed to the excessive amount of CPPS that resulted in a loose combination with the plasmid, which enlarged the nanoparticles. When the nanoparticles were prepared at a CPPS:pWnt3a weight ratio of 40:1, the particle size was significantly smaller than for the other ratio groups, which indicated that a CPPS:pWnt3a weight ratio of 40:1 might be optimal for gene transfection.
TEM was used to analyze the morphology and particle size of nanoparticles at a CPPS:pWnt3a weight ratio of 40:1. As shown in Figure 7A, most of the nanoparticles fell within a narrow range of 100–200 nm, which agreed with particle size range 167.5±6.2 nm by DLS, as Figure 7B and C show. The CPPS-Wnt3a nanoparticles prepared at a CPPS:pWnt3a weight ratio of 40:1 exhibited a uniform spherical shape, which may be conducive to cell endocytosis during transfection.52,53
In vitro transfection experiment
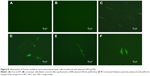
To assess transfection efficiency, CPPS-pWnt3a and CPPS-pGFP nanoparticles were prepared at three different ratios (40:1, 80:1, and 100:1). The result of pGFP transfection was analyzed by fluorescence microscopy (TI-U; Nikon, Tokyo, Japan). Lipofectamine 2000 was set as a positive control based on the manufacturer’s instructions, while HUMSCs with and without free pGFP transfection acted as negative controls. Meanwhile, the Wnt3a expression was tested by Western blotting. Lipofectamine 2000 was used as a positive control according to the manufacturer’s instructions, and the Wnt3a naked plasmid (4 μg/well) was employed as the negative control.
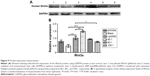
From Figure 8, it is clear that the fluorescence intensity was at the top in the 40:1 group. Meanwhile, as shown in Figure 8C, ample cell debris indicated that the cytotoxicity of the Lipofectamine 2000 group was also higher than the other groups involved, which agreed with the results of cytotoxicity assay. As Figure 9A and B demonstrate, the gene-transfection efficiency and Wnt3a expression were different for the various CPPS:pWnt3a weight ratios. The optimal Wnt3a-expression level was observed at a CPPS:pWnt3a weight ratio of 40:1. Furthermore, the transfection efficiency of the CPPS/pWnt3a nanoparticles prepared at a 40:1 weight ratio was significantly higher than that for both the 80:1 and 100:1 weight ratios (both P<0.0001, Student’s t-test). One possible reason for this result is that excess CPPS reduced the dissociation of the plasmid due to the strong bonds between the cationic carrier and the plasmid. In addition, the high cytotoxicity of a high dose of PEI could also have caused low transfection efficiency. Damage to the cell membrane caused by cationic carriers can also impact endocytosis, which is an important mechanism for gene delivery. Moreover, the transfection efficiency of Lipofectamine 2000, which was not significantly different from CPPS/pWnt3a at a weight ratio of 80:1, was obviously lower than that of CPPS/pWnt3a at a weight ratio of 40:1. This result indicated that CPPS/pWnt3a, rather than Lipofectamine 2000, could be a potential gene carrier for gene delivery.
Conclusion
In this study, we successfully created CPPS-pWnt3a nanoparticles, a novel nonviral gene vector, based on a PEI-modified polysaccharide from P. yezoensis. FTIR analyses and gel retardation assay confirmed the successful cationic modification of the original PPS. In addition, a cytotoxicity assay also showed that the 40:1 CPPS-pWnt3a nanoparticles were the least cytotoxic compared to Lipofectamine 2000 and PEI (25 kDa). The result of the cell-transfection experiment demonstrated that the optimal CPPS:pWnt3a weight ratio for gene delivery was 40:1, which was also consistent with the particle size and shape as revealed by TEM. Together, these findings indicated that CPPS could become a promising nonviral gene vector for gene delivery.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81072586, 81273470, and 81473172), the Doctoral Fund of the Ministry of Education of China (20123227110009), Special Funds for 333 Projects (BRA2013198), and industry–university–research institution cooperation (JHB2012-37, GY2012049, and GY2013055) in Jiangsu Province and Zhenjiang. This was a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Disclosure
The authors report no conflicts of interest in this work.
References
Takahashi K, Hirano Y, Araki S, Hattori M. Emulsifying ability of porphyran prepared from dried nori, Porphyra yezoensis, a red alga. J Agric Food Chem. 2000;48(7):2721–2725. | ||
Isaka S, Cho K, Nakazono S, et al. Antioxidant and anti-inflammatory activities of porphyran isolated from discolored nori (Porphyra yezoensis). Int J Biol Macromol. 2015;74:68–75. | ||
Wang J, Zhang Q, Zhang Z, Song H, Li P. Potential antioxidant and anticoagulant capacity of low molecular weight fucoidan fractions extracted from Laminaria japonica. Int J Biol Macromol. 2010;46(1):6–12. | ||
Rupérez P, Ahrazem O, Leal JA. Potential antioxidant capacity of sulfated polysaccharides from the edible marine brown seaweed Fucus vesiculosus. J Agric Food Chem. 2002;50(4):840–845. | ||
Mayer AMS, Hamann MT. Marine pharmacology in 1999: compounds with antibacterial, anticoagulant, antifungal, anthelmintic, anti-inflammatory, antiplatelet, antiprotozoal and antiviral activities affecting the cardiovascular, endocrine, immune and nervous systems, and other miscellaneous mechanisms of action. Comp Biochem Physiol C Toxicol Pharmacol. 2002;132(3):315–339. | ||
Sakai S, Komura Y, Nishimura Y, Sugawara T, Hirata T. Inhibition of mast cell degranulation by phycoerythrin and its pigment moiety phycoerythrobilin, prepared from Porphyra yezoensis. Food Sci Technol Res. 2011;17(2):171–177. | ||
Shin ES, Hwang HJ, Kim IH, Nam TJ. A glycoprotein from Porphyra yezoensis produces anti-inflammatory effects in liposaccharide-stimulated macrophages via the TLR4 signaling pathway. Int J Mol Med. 2011;28(5):809–815. | ||
Cao J, Wang S, Yao C, Xu Z, Xu X. Hypolipidemic effect of porphyran extracted from Pyropia yezoensis in ICR mice with high fatty diet. J Appl Phycol. Epub 2015 Jun 5. | ||
Kitajima H, Komizu Y, Ichihara H, et al. Immunostimulatory effects of extract from nori (Porphyra yezoensis) in vitro and in vivo. Kagaku Kogaku Ronbunshu. 2013;39(4):359–362. | ||
Kwon MJ, Nam TJ. Chromatographically purified porphyran from Porphyra yezoensis effectively inhibits proliferation of human cancer cells. Food Sci Biotechnol. 2007;16(6):873–878. | ||
Park SJ, Ryu J, Kim IH, Choi YH, Nam TJ. Activation of the mTOR signaling pathway in breast cancer MCF627 cells by a peptide derived from Porphyra yezoensis. Oncol Rep. 2015;33(1):19–24. | ||
Klink D, Schindelhauer D, Laner A, et al. Gene delivery systems – gene therapy vectors for cystic fibrosis. J Cyst Fibros. 2004;3 Suppl 2:203–212. | ||
Han S, Mahato RI, Sung YK, Kim SW. Development of biomaterials for gene therapy. Mol Ther. 2000;2(4):302–317. | ||
de Carvalho Rodrigues D, Asensi KD, Vairo L, et al. Human menstrual blood-derived mesenchymal cells as a cell source of rapid and efficient nuclear reprogramming. Cell Transplant. 2012;21(10):2215–2224. | ||
Fujikura J, Nakao K, Sone M, et al. Induced pluripotent stem cells generated from diabetic patients with mitochondrial DNA A3243G mutation. Diabetologia. 2012;55(6):1689–1698. | ||
Kane NM, Nowrouzi A, Mukherjee S, et al. Lentivirus-mediated reprogramming of somatic cells in the absence of transgenic transcription factors. Mol Ther. 2010;18(12):2139–2145. | ||
Shao L, Feng W, Sun Y, et al. Generation of iPS cells using defined factors linked via the self-cleaving 2A sequences in a single open reading frame. Cell Res. 2009;19(3):296–306. | ||
Wang WW, Wei W, Yan J, et al. Reprogramming of mouse renal tubular epithelial cells to induced pluripotent stem cells. Cytotherapy. 2013;15(5):578–585. | ||
Moriguchi H, Chung RT, Mihara M, Sato C. Generation of human induced pluripotent stem cells from liver progenitor cells by only small molecules. Hepatology. 2010;52(3):1169. | ||
Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(3):861–872. | ||
Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. | ||
Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. | ||
Choi JW, Jung SJ, Kasala D, et al. pH-sensitive oncolytic adenovirus hybrid targeting acidic tumor microenvironment and angiogenesis. J Control Release. 2015;205:134–143. | ||
Lee CH, Kim HW, Kim T, Lee SW. Recombinant adenovirus infection suppresses hTERT expression through virus-associated RNA-mediated induction of type 1 interferon. Biochem Biophys Res Commun. 2015;458(4):830–835. | ||
Rodríguez-García A, Giménez-Alejandre M, Rojas JJ, et al. Safety and efficacy of VCN-01, an oncolytic adenovirus combining fiber HSG-binding domain replacement with RGD and hyaluronidase expression. Clin Cancer Res. 2015;21(6):1406–1418. | ||
Pendaries V, Gasc G, Titeux M, et al. Immune reactivity to type VII collagen: implications for gene therapy of recessive dystrophic epidermolysis bullosa. Gene Ther. 2010;17(7):930–937. | ||
Mcmenamin MM, Wood MJ. Progress and prospects: immunobiology of gene therapy for neurodegenerative disease: prospects and risks. Gene Ther. 2010;17(4):448–458. | ||
Svahn MG, Lundin KE, Ge R, et al. Adding functional entities to plasmids. J Gene Med. 2004;6 Suppl 1:S36–S44. | ||
Bacharach E, Gonsky J, Lim D, Goff SP. Deletion of a short, untranslated region adjacent to the polypurine tract in Moloney murine leukemia virus leads to formation of aberrant 5´ plus-strand DNA ends in vivo. J Virol. 2000;74(10):4755–4764. | ||
Somanathan S. AAV vectors avoid inflammatory signals necessary to render transduced hepatocyte targets for destructive T cells. Mol Ther. 2010;18(5):977–982. | ||
Atkinson H, Chalmers R. Delivering the goods: viral and non-viral gene therapy systems and the inherent limits on cargo DNA and internal sequences. Genetica. 2010;138(5):485–498. | ||
Yu G, Borlongan CV, Ou Y, et al. In vitro non-viral Lipofectamine delivery of the gene for glial cell line-derived neurotrophic factor to human umbilical cord blood CD34+ cells. Brain Res. 2010;1325(1):147–154. | ||
Gadi J, Ruthala K, Kong KA, Park HW, Kim MH. The third helix of the Hoxc8 homeodomain peptide enhances the efficiency of gene transfer in combination with Lipofectamine. Mol Biotechnol. 2009;42(1):41–48. | ||
Kim HK, Davaa E, Myung CS, Park JS. Enhanced siRNA delivery using cationic liposomes with new polyarginine-conjugated PEG-lipid. Int J Pharm. 2010;392(1–2):141–147. | ||
Davaa E, Kang BS, Han JH, et al. Combined delivery of the adiponectin gene and rosiglitazone using cationic lipid emulsions. Int J Pharm. 2015;483(1–2):124–130. | ||
Tanasienko IV, Yemets AI, Finiuk NS, Stoika RR, Blume YB. DMAEM-based cationic polymers as novel carriers for DNA delivery into cells. Cell Biol Int. 2015;39(3):243–245. | ||
Slita AV, Kasyanenko NA, Nazarova OV, et al. DNA-polycation complexes: effect of polycation structure on physico-chemical and biological properties. J Biotechnol. 2007;127(4):679–693. | ||
Filion MC, Phillips NC. Major limitations in the use of cationic liposomes for DNA delivery. Int J Pharm. 1998;162(1–2):159–170. | ||
Deng WW, Cao X, Wang M, et al. Efficient gene delivery to mesenchymal stem cells by an ethylenediamine-modified polysaccharide from mulberry leaves. Small. 2012;8(3):441–451. | ||
Wang M, Deng W, Fu M, et al. Efficient gene transfer into rat mesenchymal stem cells with cationized Lycium barbarum polysaccharides nanoparticles. Carbohydr Polym. 2011;86(4):1509–1518. | ||
Deng W, Fu M, Cao Y, et al. Angelica sinensis polysaccharide nanoparticles as novel non-viral carriers for gene delivery to mesenchymal stem cells. Nanomedicine. 2013;9(8):1181–1191. | ||
Deng WW, Cao X, Wang M, et al. Delivery of a transforming growth factor β-1 plasmid to mesenchymal stem cells via cationized Pleurotus eryngii polysaccharide nanoparticles. Int J Nanomedicine. 2012;7:1297–1311. | ||
Cao X, Deng W, Qu R, et al. Non-viral co-delivery of the four Yamanaka factors for generation of human induced pluripotent stem cells via calcium phosphate nanocomposite particles. Adv Funct Mater. 2013;23(43):5403–5411. | ||
Yang XT, Bi YY, Chen ET, Feng DF. Overexpression of Wnt3a facilitates the proliferation and neural differentiation of neural stem cells in vitro and after transplantation into an injured rat retina. J Neurosci Res. 2014;92(2):148–161. | ||
Davidson KC, Jamshidi P, Daly R, Hearn MT, Pera MF, Dottori M. Wnt3a regulates survival, expansion, and maintenance of neural progenitors derived from human embryonic stem cells. Mol Cell Neurosci. 2007;36(3):408–415. | ||
Lee-Kubli CA, Lu P. Induced pluripotent stem cell-derived neural stem cell therapies for spinal cord injury. Neural Regen Res. 2015;10(1):10–16. | ||
Li X, Saiki C, Ide R. Stem cell therapy for central nerve system injuries: glial cells hold the key. Neural Regen Res. 2014;9(13):1253–1260. | ||
Nicolas G, Franklin RJ, Jeffery ND. Cell therapy for spinal cord injuries: what is really going on? Neuroscientist. 2014;20(6):623–638. | ||
Matsui T, Akamatsu W, Nakamura M, Okano H. Regeneration of the damaged central nervous system through reprogramming technology: basic concepts and potential application for cell replacement therapy. Exp Neurol. 2014;260(5):12–18. | ||
Xu X, Capito RM, Spector M. Delivery of plasmid IGF-1 to chondrocytes via cationized gelatin nanoparticles. J Biomed Mater Res A. 2008;84(1):73–83. | ||
Yang HJ, Pei F, Lei W, et al. Caveolin-1 mediates gene transfer and cytotoxicity of polyethyleneimine in mammalian cell lines. Mol Cell Biochem. 2015;402(1–2):203–211. | ||
Xu X, Capito RM, Spector M. Plasmid size influences chitosan nanoparticle mediated gene transfer to chondrocytes. J Biomed Mater Res A. 2008;84A(4):1038–1048. | ||
MacLaughlin FC, Mumper RJ, Wang J, et al. Chitosan and depolymerized chitosan oligomers as condensing carriers for in vivo plasmid delivery. J Control Release. 1998;56(1–3):259–272. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.