Back to Journals » Neuropsychiatric Disease and Treatment » Volume 18
Effect of the Minor C Allele of CNTN4 rs2619566 on Medial Hypothalamic Connectivity in Early-Stage Patients of Chinese Han Ancestry with Sporadic Amyotrophic Lateral Sclerosis
Authors Wang Y , He Y, Zhu Y , He T, Xu J, Kuang Q, Ji Y, Xu R, Li F, Zhou F
Received 16 September 2021
Accepted for publication 1 February 2022
Published 25 February 2022 Volume 2022:18 Pages 437—448
DOI https://doi.org/10.2147/NDT.S339456
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yuping Ning
Yao Wang,1,2 Yujie He,1,2 Yanyan Zhu,1,2 Ting He,1,2 Jie Xu,1,2 Qinmei Kuang,1,2 Yuqi Ji,1,2 Renshi Xu,3 Fangjun Li,4 Fuqing Zhou1,2
1Department of Radiology, The First Affiliated Hospital of Nanchang University, Nanchang, 330006, People’s Republic of China; 2Neuroimaging Lab, Jiangxi Province Medical Imaging Research Institute, Nanchang, 330006, People’s Republic of China; 3Department of Neurology, Jiangxi Provincial People’s Hospital Affiliated to Nanchang University, Nanchang, 330006, People’s Republic of China; 4Department of Neurology, The First Affiliated Hospital of Nanchang University, Nanchang, 330006, People’s Republic of China
Correspondence: Fuqing Zhou, Department of Radiology, The First Affiliated Hospital of Nanchang University, No. 17 Yongwaizheng Street, Nanchang, 330006, People’s Republic of China, Tel +86- 13694881033, Email [email protected]; [email protected] Fangjun Li, Department of Neurology, The First Affiliated Hospital of Nanchang University, No. 17 Yongwaizheng Street, Nanchang, 330006, People’s Republic of China, Tel +86- 13576937919, Email [email protected]
Objective: Clinical symptoms such as major defects in energy metabolism may involve the hypothalamus in amyotrophic lateral sclerosis (ALS) patients. Our recent study discovered that the single-nucleotide polymorphisms (SNPs) of rs2619566, rs79609816 and rs10260404 are associated with sporadic ALS (sALS). Thus, this study aims to investigate the hypothalamic functional reorganization and its association with the above polymorphisms risk alleles in sALS patients of Chinese Han ancestry.
Methods: Forty-four sALS patients (28 males/16 females) and 40 healthy subjects (HS; 28 males/12 females) underwent resting-state functional MRI, genotyping and clinical assessments. A two-sample t test (P < 0.01, GRF correction at P < 0.05) was performed to compare hypothalamic connectivity for group-level analysis in disease diagnosis and genotype, and then the genotype-diagnosis interaction effect was assessed. Finally, Spearman correlation analyses were performed to assess the relationship between the altered functional connectivity and their clinical characteristics.
Results: The sALS patients showed a short disease duration (median = 12 months). Regarding the diagnosis effect, the sALS patients showed widespread hypothalamic hyperconnectivity with the left superior temporal gyrus/middle temporal gyrus, right inferior frontal gyrus, and left precuneus/posterior cingulate gyrus. For the genotype effect of SNPs, hyperconnectivity was observed in only the medial hypothalamus when the sALS patients harboring the minor C allele of rs2619566 in contactin-4 (CNTN4), while the sALS patients with the TT allele showed a hyperconnectivity network in the right lateral hypothalamus. This connectivity pattern was not observed in other SNPs. No significant genotype-diagnosis interaction was found. Moreover, altered functional connectivity was not significantly correlated with clinical characteristics (P : 0.11– 0.90).
Conclusion: These results demonstrated widespread hypothalamic hyperconnectivity in sALS. The risk allele C of the CNTN4 gene may therefore influence functional reorganization of the medial hypothalamus. The effects of the CNTN4 rs2619566 polymorphism may exist in the hypothalamic functional connectivity of patients with sALS.
Keywords: sporadic amyotrophic lateral sclerosis, hypothalamus, lateral hypothalamus, medial hypothalamus, resting-state functional connectivity
Corrigendum for this paper has been published
Introduction
Amyotrophic lateral sclerosis (ALS) is a neuromuscular disorder characterized by progressive muscle atrophy and paralysis, in which the affected individual eventually dies of breathing failure. ALS is characterized by both upper and lower motor neuron dysfunction, but is not limited to these regions, eg, structural or functional impairment in the middle temporal gyrus,1 insula,2,3 anterior cingulate gyrus4 and some regions within the occipital and parietal lobes.5
As an integration of the periphery and central nervous system, the hypothalamus governs many physiological processes such as thermoregulation, energy balance, circadian rhythms, and reproductive behavior. More recent studies have suggested that functional and structural impairments within the hypothalamus may be involved in patients with ALS.6,7 For example, ALS patients may suffer from major defects in energy metabolism due to losing control of the hypothalamic melanocortin pathway.6 Neuroimaging studies have observed that hypothalamus atrophy in ALS in the premorbid stage and correlates with body mass index (BMI).8 Furthermore, a diffusion tensor imaging study demonstrated the selective involvement of orbitofrontal, insular, and motor cortex projections to the hypothalamus in murine ALS models and human patients,3 and some studies showed that neurons of the hypothalamus innervate various subcortical motor structures such as the cerebellum, vestibular nuclei9 and even spinal cord,10 which may be associated with motor dysfunction in ALS.
Recent research suggests that genetic factors may also play an important role in sporadic ALS (sALS). Our recent study discovered that subjects harboring the minor C allele of the rs2619566 polymorphism in the contactin-4 (CNTN4) gene and the minor T allele of rs79609816 in the inositol polyphosphate-5-phosphatase B (INPP5B) gene exhibited an increased risk for sALS development, but the minor C allele of rs10260404 in the dipeptidyl-peptidase 6 (DPP6) played a protective role in Chinese Han ancestry.11 Comparatively, the mechanism of CNTN4 in ALS disease has been more recognized. CNTN4 gene encodes neuron adhesion molecules,12 and harboring the minor C allele (CC + CT) induces abnormalities in differentiation, adhesion, projection, and synaptic plasticity of motor neurons, disturbances in neuronal network formation and plasticity of sALS.11
Thus, in this study, we hypothesized that functional disruption and/or plasticity might occur in large-scale hypothalamic functional networks and that there are differences in sALS patients with different genotypes of SNPs. To achieve this goal, we first employed resting-state functional connectivity (FC), a reliable and widely used technique, to investigate the alteration of the hypothalamic functional network in the hypothalamus and its subregions, the medial hypothalamus (MH) and the lateral hypothalamus (LH), which involve in waking/feeding and resting/satiety, respectively. Second, we investigated the patterns of functional connectivity in the different genotype groups and assessed the hypothalamic connectivity as a possible marker of disease severity in sALS.
Subjects and Methods
Subjects
A total of 60 patients with sALS were recruited from our hospital from April 2013 to December 2018. All patients were diagnosed according to the revised El Escorial criteria. To estimate functional status in sALS, an “ALS Functional Rating Scale (ALSFRS)” was performed on each sALS subject within 2 hours before the MRI scans. A lower ALSFRS score indicates more severe neurological dysfunction in sALS patients. The patients were not considered to have a family history of motor neuron disease, psychiatric disorder, or other nervous system illnesses, including clinically diagnosed frontotemporal dementia. Fifty-one healthy subjects (HS) were enrolled in this investigation.
All participants provided written informed consent following the Helsinki Declaration and approved guidelines set by the Human Research Ethics Committee and our hospital’s Institutional Review Board.
Selection of SNPs
This study was based on our previous researches: First, 7 SNPs and 20 possible loci associated with sALS were selected based on a genome-wide association study and other studies.11,13 Then, the abovementioned 27 SNPs were analyzed for 239 sALS cases and 261 controls to further explore possible risk gene loci in Chinese Han ancestry and reported that the polymorphisms rs2619566 in CNTN4, rs10260404 in DPP6, and rs79609816 in INPP5B were defined as possible genetic risk factors and may play roles in the pathogenesis of sALS.11 Thus, we selected the abovementioned 3 SNPs for further genotyping.
Genotyping
Genomic DNA from peripheral blood containing ethylenediaminetetraacetic acid (EDTA) was extracted using standard techniques. For the genotyping of each SNP, the primers CNTN4 rs2619566, DPP6 rs10260404, and INPP5B rs79609816 were designed using Primer3 online software (http://primer3.wi.mit.edu/). Multiplex polymerase chain reactions were performed to amplify the target SNPs. For PCR products, Sanger sequencing was performed on an ABI 3730XL DNA analyzer (Applied Biosystems, Inc. CA) to identify the genotyping results of the 3 SNPs. Then, the sequencing results were analyzed with SeqMan software (DNASTAR, Lasergene) and compared with the reference sequence obtained from NCBI GenBank for mutation analysis.
MRI Acquisition
A 3T SIEMENS scanner (Trio, Siemens, Munich, Germany) was utilized to obtain all imaging data in the Department of Radiology at our hospital. During data collection, each subject was required to close their eyes, remain awake and avoid systematic thinking. Subjects were also required to remain motionless as much as possible. The resting-state functional MRI (rs-fMRI) data were acquired with an echo-planar imaging (EPI) sequence using the following settings: repetition time/echo time (TR/TE): 2000 ms/30 ms, flip angle: 90°, slices: 30, thickness: 4 mm, field of view (FOV): 220 mm × 220 mm, in-plane matrix: 64 × 64; The parameters of a high-resolution and three-dimensional T1-weighted sequence were as follows: TR: 1900 ms, TE: 2.26 ms, flip angle: 15°, FOV: 215 mm × 230 mm, matrix size: 240 × 256, 176 slices.
Data Preprocessing
For each subject, the preprocessing of rs-fMRI was performed by Data Processing & Analysis of Brain Imaging (DPABI_3.0, http://rfmri.org/dpabi) and a statistical parametric mapping (SPM12, http://www.fil.ion.ucl.ac.uk/spm/software/spm12/) software package, which is based on the MATLAB 8.4.0 (The MathWorks Inc., Natick, MA, USA). The main steps of data preprocessing included the followings: 1) The first 10 volumes were removed to maintain brain signals equilibrium; 2) The remaining volumes then underwent slices time correction and head motion realignment. The participants’ images were excluded if their head motion exceeded 2mm translation or 2° rotation. Microhead movement (mean frame displacement, mean FD) was measured for each subject according to the result of Van Dijk et al;14 3) Functional images were registered to T1 weighted images, and T1 image segmentations and functional images were normalized to Montreal Neurological Institute (MNI) space with resampling to 3×3×3 mm3; 4) After the normalization described above, the nuisance linear regression was analyzed with the average blood oxygenation level dependent signals of cerebrospinal fluid, white matter, and 24 head motion parameters as covariates; 5) Finally, a 0.01–0.10 Hz band-pass filtration was applied for resting-state functional MRI data to reduce drift and physiological noise.
Functional Connectivity and Group Analyses
Seed-based functional connectivity was utilized to examine alterations in hypothalamic FC in patients with ALS. First, the peak MNI coordinate of hypothalamic seed was selected at x = 2, y = −1, z = −12, with a radius of 4 mm. Furthermore, according to previous studies,15,16 two paired spherical seeds were defined with a radius of 2 mm, with the coordinates of the MH and LH (MH: ±4, −2, −12; LH: ±6, −9, −10) as the centers. The average blood oxygenation level-dependent time series for each seed was extracted separately as a reference time course, and then we applied the Pearson correlation to quantify the relationship between these seeds and the remaining brain regions at the voxel-based level. Five connectivity maps were generated, and then Fisher’s Z transformation of each subject was performed for group analysis.
Statistical Analysis
Statistical analyses of the demographic and clinical characteristics were evaluated in SPSS 23.0 (Chicago, IL, USA), and the Kolmogorov–Smirnov test was used to check for the normality among all data above. Hardy–Weinberg equilibrium was assessed in sALS and HS. Functional connectivity was estimated within the following groups: (1) diagnosis group (sALS vs HS); (2) genotype subgroups of 3 SNPs; (3) genotype-diagnosis interaction effect. For not normally distributed variables, median and range were used to describe the trend of variables, while mean and standard deviation (SD) were for normally distributed measures of interest. The chi-square test was utilized to compare sex differences within groups, whereas a two-sample t test was for age and mean FD. A two-sample t test (P < 0.01) analysed group differences to investigate functional connectivity network of the hypothalamus, MH and LH, age, sex, and mean FD as covariates. Gaussian random field theory (GRF) correction was performed for multiple comparisons (P < 0.05). Since clinical variables were not normally distributed, we chose Spearman correlation to measure the association between the altered hypothalamic functional index and clinical characteristics (sex, age and mean FD as covariates).
Results
Participants
In this study, 27 subjects were excluded due to poor quality MRI, incomplete clinical data, and intracranial lesions. Ultimately, 44 sALS patients and 40 HS were included for further analysis in this study. The distributions of genotypes of the 3 SNPs are shown in Table 1. According to the genotypes of 3 SNPs, the sALS and HS groups were divided into 2 subgroups for more functional connectivity comparisons (Table 1). No significant differences in age (P = 0.869), gender (P = 0.537) and mean FD (P = 0.334) between sALS and HS groups (Table 2). A short disease duration (median = 12 months) indicated that the sALS patients were in the early stage of disease.
 |
Table 1 Summary of Single-Nucleotide Polymorphisms Identified Across Analyses |
 |
Table 2 Groups and Clinical Characteristics of the sALS Group and CNTN4 Subgroups |
Altered Hypothalamic FC Network in sALS and Genotype Subgroups
Compared with HS, we observed increased hypothalamic FC in the sALS patients, which mainly distributed in the left superior temporal gyrus/middle temporal gyrus, right inferior frontal gyrus, and left precuneus/posterior cingulate gyrus (Figure 1 and Table 3). Similar hyperconnectivity was also observed in the sALSC of CNTN4 subgroup (Figure 1 and Table 3), the sALSA of INPP5B subgroup (Supplementary Material; Table S1), and the sALST of DPP6 subgroup (Supplementary Material; Table S2). However, compared with the SNPs subgroup of HS, the sALST of CNTN4, sALSC of DPP6 and sALST of INPP5B subgroups did not show significant differences on hypothalamic FC. No significant genotype-diagnosis interaction was found in hypothalamic functional connectivity.
 |
Table 3 Altered Brain Regions of the Hypothalamic Network in the sALS Group and sALSC of CNTN4 Subgroup |
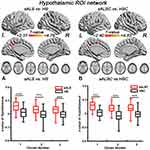 |
Figure 1 Brain regions with significantly increased hypothalamic FC in sALS group (A) and sALSC of the CNTN4 subgroup (B) (P < 0.01, GRF correction at P < 0.05), the corresponding box diagrams are located at the bottom, respectively. Abbreviations: ROI, region of interest; sALS, sporadic amyotrophic lateral sclerosis; HS, healthy subjects; sALSC, sALS carrying the minor C allele of CNTN4 rs2619566; HSC, HS carrying the minor C allele of CNTN4 rs2619566; L, left; R, right. Notes: ***Means P < 0.001. Details of cluster number are shown in Table 3. |
Comparison of FC in the Hypothalamic Subregions (MH and LH)
In the MH functional network, sALS patients showed increased FC between the MH and several regions compared with HS, including the left middle temporal gyrus, left posterior cingulate/cuneus, bilateral precuneus, and cerebellum; similar hyperconnectivity of the MH was also observed in the sALSC of CNTN4 subgroup (for details, see Figure 2 and Table 4). For the LH functional network, we observed only increased right LH FC with some brain regions in the occipital lobe in sALS groups; sALST of CNTN4 subgroup also showed similar hyperconnectivity of the right LH (for details see Figure 3 and Table 5).
 |
Table 4 Altered Brain Regions of the MH Network in the sALS Group and sALSC of CNTN4 Subgroup |
 |
Table 5 Altered Brain Regions of the Right LH Network in the sALS Group and sALST of CNTN4 Subgroup |
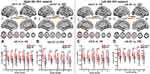 |
Figure 2 Brain regions with significantly increased FC of the right MH (A and B) and left MH (C and D) in the sALS group and sALSC of CNTN4 subgroups (P < 0.01, GRF correction at P < 0.05), the corresponding box diagrams are located at the bottom, respectively. Abbreviations: MH, medial hypothalamus; ROI, region of interest; sALS, sporadic amyotrophic lateral sclerosis; HS, healthy subjects; sALSC, sALS carrying the minor C allele of CNTN4 rs2619566; HSC, HS carrying the minor C allele of CNTN4 rs2619566; L, left; R, right. Notes: ***Means P < 0.001. Details of cluster number are shown in Table 4. |
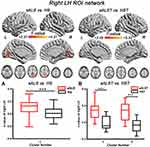 |
Figure 3 Brain regions with significantly increased right LH FC in the sALS group (A) and sALST of CNTN4 subgroup (B) (P < 0.01, GRF correction at P < 0.05), the corresponding box diagrams are located at the bottom, respectively. Abbreviations: LH, lateral hypothalamus; ROI, region of interest; sALS, sporadic amyotrophic lateral sclerosis; HS, healthy subjects; sALST, sALS carrying the TT allele of CNTN4 rs2619566; HST, HS carrying the TT allele of CNTN4 rs2619566; L, left; R, right. Notes: ***Means P < 0.001. Details of cluster number are shown in Table 5. |
For the DPP6 and INPP5B genes, hyperconnectivity occurred in the bilateral MH or LH ROI of the sALST of DPP6 and sALSA of INPP5B subgroups, similar to the hypothalamic FC network. However, no alteration of FC was detected in the sALSC of DPP6 or sALST of INPP5B subgroups. Detailed results are reported in Tables S1 and S2. No significant genotype-diagnosis interaction was observed in MH and LH functional connectivity.
Correlation Analysis
Interestingly, we found that the altered FC of the hypothalamus (including MH and LH) was not related to patients’ physical ability (ALSFRS score) within ALS and CNTN4 subgroups (detail showed in Supplementary Material; Table S3).
Discussion
In this study, we observed hypothalamic hyperconnectivity in patients with sALS, indicating a wide functional organization of the hypothalamus. When the genotype effects of SNPs were assessed, we observed that the sALSC of CNTN4 subgroup showed hyperconnectivity only in the MH functional network rather than LH. Correspondingly, the sALST of CNTN4 subgroup exhibited increased FC in the LH functional network. In addition, hyperconnectivity was found only in the major A or T alleles of the DPP6 and INPP5B genes.
Hypothalamic Functional Hyperconnectivity in sALS Patients
In addition to progressive degeneration of motor neurons, ALS patients are well known to suffer from energy metabolic dysfunction (eg, weight loss) and eating behavior changes.17,18 Previous pathological studies have shown that weight loss in ALS may be related to hypothalamic atrophy or pathological deposition of TDP-43 in the hypothalamus,7,8 suggesting that the hypothalamus plays a key role in the energy metabolism of ALS patients. In this study, we observed the hyperconnectivity between hypothalamus and left superior temporal gyrus/middle temporal gyrus in sALS patients. Earlier evidence has reported that neural activation in the superior temporal gyrus/middle temporal gyrus was positively associated with weight loss.19 Moreover, a recent study on eating behavior-induced brain structure changes showed that grey matter volume (GVM) in the middle temporal gyrus is associated with cognitive restraint of eating scores and BMI; this study also demonstrated that GVM in the hypothalamus has a negative correlation with hunger score.20 According to previous works, our findings may exhibit functional connectivity alterations between brain regions in response to changes in energy metabolism and food intake in ALS. Similar to the reports of abnormal functional connectivity and cortical damage that have been reported in the precuneus/posterior cingulate gyrus in ALS patients,21,22 our results also showed increased hypothalamic FC with the left precuneus/posterior cingulate gyrus in ALS. The precuneus/posterior cingulate gyrus is regarded as an essential part of the default mode network (DMN), which is described as being strongly associated with aging,23 cognitive deficits,24 higher disability, and faster progression rate in ALS.25 In keeping with the above research, our results may reflect ALS-related changes in cognitive status and behavior changes.2,26 Furthermore, increased FC between hypothalamus and right lentiform nucleus/putamen was also observed in ALS, which may suggest subcortical network reorganization of sALS patients.26
Functional Connectivity in the Sub-Regions (MH and LH)
When the hypothalamus was divided into MH and LH, in addition to the brain regions above, a greater range of hyperconnectivity was observed in sALS and its subgroups. In the MH functional network, the cerebellum was included in the sALS groups. The cerebellum integrates sensory and motor inputs, which plays a significant role in motor control and modulate cognitive/emotional functions, while MH is involved in attack and defense system.27,28 Animal mode has found the axonal projections from dorsomedial hypothalamus (an essential component of medial hypothalamus) to cerebellum,29 and our findings support the results of functional MRI and indicate that MH may also be involved in the regulation of cerebellar functions in sALS patients.
Interestingly, in the LH functional network, sALST patients exhibited increased right LH FC with visual areas in the occipital lobe as similar to sALS group. Tigges et al reported projections from the LH to the parietal and occipital cortices in chimpanzee modes.30 In humans, Kamali et al also demonstrated the long direct connectivity from hypothalamus to the parietal and occipital lobes by diffusion-weighted imaging,31 which may carry the visuosensory-related information directly from the visuosensory cortices to the limbic system. Our results may be strongly associated with this increased transmission of visuosensory information in sALS.
The Minor C Allele of CNTN4 rs2619566 is Associated with Medial Hypothalamic Functional Connectivity in Patients with sALS
When evaluating the genotype effect of SNPs in this study, We found that sALS patients harboring the minor C allele (risk SNPs) of CNTN4 rs2619566 showed hyperconnectivity in the MH, but this pattern of alteration was not seen with other risk SNPs, eg, the minor C allele of DPP6 rs10260404 and the minor T allele of INPP5B rs79609816. A previous study reported that CNTN4 was closely associated with sALS (P < 0.01) as a high-risk factor for sALS.13 Carriers of the minor C allele of CNTN4 rs2619566 may damage neuronal axons and influence neuronal differentiation, cell–cell adhesion, neuronal projection, and synaptic plasticity. Neural network damage and altered plasticity may subsequently induce sALS development.11 Our results indicated that the minor C allele of rs2619566 may influence the formation and plasticity of the MH functional network in sALS patients, as disruptions of CNTN4 expression were reported to potentially lead to developmental delay or defects.32 CNTN4 is an autism susceptibility locus,33 and CNTN4 is the most widely expressed in the thalamus, including most of the thalamic nucleus,34 while in a mouse model, CNTN4 mutant autism mice showed reduced cell surface glutamate receptors in the cortex and hippocampus and increased cell surface glutamate receptors in the striatum.35 Moreover, widespread hyperconnectivity between MH and the bilateral lentiform nucleus/putamen was found in sALS patients with the minor C allele of CNTN4. These findings indicate that the CNTN4 variant may disturb the formation and plasticity of the MH functional network by changing the levels of expression of some cell surface receptors in patients with sALS. Indeed, we assessed the effect of the CNTN4 rs2619566 SNP on grey matter volume, the white matter fibre bundle, the intensity of spontaneous activity in brain regions, and resting state functional network connectivity in sALS patients (manuscript in preparation). We indeed found that sALS patients carrying minor C allele showed decreased grey matter density in the right parahippocampal gyrus and subcortical nucleus, and extensive white matter fibre damage. Increased functional connectivity was thought to be a compensatory mechanism for structural damage, which further supports our findings that widespread hypothalamic hyperconnectivity occurs in sALS and sALSC in the CNTN4 rs2619566 groups. Using independent component analysis, functional connectivity network analysis suggested increased functional connectivity in the right lentiform nucleus, which positively correlated with ALSFRS-R scores. Combined with negative findings on interactions, our results may indicate that the minor C allele of CNTN4 rs2619566 may influence the formation and plasticity of the MH functional network in sALS patients. However, the mechanism and clinical significance of CNTN4-related hyperconnectivity in MH is still unknown, and further studies are needed in the future.
Limitations
Although our findings are novel, this study still has some limitations. First, our sample was small size due to the low morbidity of sALS in China. Thus, studies with large sample sizes will be conducted in our future works to further support these findings. Second, as a retrospective study, only ALSFR scores were utilized to estimate sALS patients’ physical disability. Some patients with sALS have cognitive impairments, sleep disorders, and weight changes, which may be closely associated with the hypothalamus. The lack of specific neuropsychological assessments might limit our interpretation of the correlation analysis. Finally, there is a lack of screening for mutations causing familial ALS since the current study predominantly focuses on patients with sporadic ALS. Therefore, we should perform a study considering the above points in further future.
Conclusion
Extensive hyperconnectivity of the hypothalamus was observed in early-stage patients with sALS, demonstrating the underlying hypothalamic functional reorganization. Moreover, early-stage patients harboring the minor C allele of CNTN4 rs2619566 may be associated with medial hypothalamic functional connectivity in sALS. It would be interesting to explore the relationship between CNTN4 rs2619566 and hypothalamic functional connectivity with clinical parameters (eg, hunger, weight loss, and anorexia) in the future for a deeper understanding of the hypothalamic functional alterations in sALS.
Abbreviations
sALS, sporadic amyotrophic lateral sclerosis; CNTN4, contactin-4; HS, healthy subjects; DPP6, dipeptidyl-peptidase 6; INPP5B, inositol polyphosphate-5-phosphatase B; Chr, chromosome; GRF, Gaussian random fields; FC, functional connectivity; MH, medial hypothalamus; LH, lateral hypothalamus; ROI, region of interest; TDP-43, transactive response DNA-binding protein 43 kDa; BMI, body mass index; SNP, single nucleotide polymorphism; rs-fMRI, resting-state functional MRI; EPI, echo-planar imaging; TR, repetition time; TE, echo time; FOV, field of view; DPABI, Data Processing & Analysis of Brain Imaging; FD, frame displacement; MNI, Montreal Neurological Institute; SD, standard deviation; ALSFRS, ALS Functional Rating Scale; N.A, not applicable; L, left; R, right; GVM, grey matter volume; DMN, default mode network.
Ethics Approval and Informed Consent
All procedures carried out in the study were in accordance with the ethical standards approved by the Medical Ethics Committee of the First Affiliated Hospital of Nanchang University [Grant number: 4-074]. The subjects gave written informed consent prior to participation.
Consent for Publication
We have obtained the informed consent from all patients or their legal guardians.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China [Grant numbers: 82160331 and 81771808], the Jiangxi Provincial Key Research and Development Program [Grant number: 20192BBGL70034], the Jiangxi provincial Department of science and technology [Grant number: 20171BAB215022], the General Projects of Provincial Traditional Chinese Medicine Science and Technology Plan [Grant number: 2020B0336], and the Science and Technology Planning Project of Jiangxi Provincial Health Commission [Grant number: 202110014].
Author Contributions
All authors made substantial contributions to conception, study design, execution, acquisition of data, or analysis and interpretation of data; have taken part in drafting the article or revising it critically for some important sections of the work; agreed to submit to the current journal; gave approval of the final version to be published; and agree to be accountable for the contents of the article.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bueno APA, Pinaya WHL, Rebello K, et al. Regional dynamics of the resting brain in amyotrophic lateral sclerosis using fractional amplitude of low-frequency fluctuations and regional homogeneity analyses. Brain Connect. 2019;9(4):356–364. doi:10.1089/brain.2019.0663
2. Trojsi F, Di Nardo F, Siciliano M, et al. Resting state functional MRI brain signatures of fast disease progression in amyotrophic lateral sclerosis: a retrospective study. Amyotroph Lateral Scler Frontotemporal Degener. 2021;22:117–126. doi:10.1080/21678421.2020.1813306
3. Bayer D, Antonucci S, Müller H-P, et al. Orbitofrontal-hypothalamic projections are disrupted in hypermetabolic murine ALS model and human patients. Transl Neurodegener. 2021;10:1–7. doi:10.1101/2020.11.28.402065
4. Braak H, Del Tredici K. Anterior cingulate cortex TDP-43 pathology in sporadic amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2018;77(1):74–83. doi:10.1093/jnen/nlx104
5. Agosta F, Valsasina P, Riva N, et al. The cortical signature of amyotrophic lateral sclerosis. PLoS One. 2012;7(8):e42816. doi:10.1371/journal.pone.0042816
6. Vercruysse P, Sinniger J, El Oussini H, et al. Alterations in the hypothalamic melanocortin pathway in amyotrophic lateral sclerosis. Brain. 2016;139(Pt 4):1106–1122. doi:10.1093/brain/aww004
7. Cykowski MD, Takei H, Schulz PE, Appel SH, Powell SZ. TDP-43 pathology in the basal forebrain and hypothalamus of patients with amyotrophic lateral sclerosis. Acta Neuropathol Commun. 2014;2(1):1–11. doi:10.1186/s40478-014-0171-1
8. Gorges M, Vercruysse P, Muller HP, et al. Hypothalamic atrophy is related to body mass index and age at onset in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2017;88(12):1033–1041. doi:10.1136/jnnp-2017-315795
9. Zhang XY, Yu L, Zhuang QX, et al. Hypothalamic histaminergic and orexinergic modulation on cerebellar and vestibular motor control. Cerebellum. 2013;12(3):294–296. doi:10.1007/s12311-012-0442-y
10. Wu G-Y, Han X-H, Zhuang Q-X, et al. Excitatory effect of histamine on rat spinal motoneurons by activation of both H1 and H2 receptors in vitro. J Neurosci Res. 2012;90(1):132–142. doi:10.1002/jnr.22730
11. Zhang J, Qiu W, Hu F, et al. The rs2619566, rs10260404, and rs79609816 polymorphisms are associated with sporadic amyotrophic lateral sclerosis in individuals of Han Ancestry From Mainland China. Front Genet. 2021;12:679204. doi:10.3389/fgene.2021.679204
12. Zeng L, Zhang C, Xu J, et al. A novel splice variant of the cell adhesion molecule contactin 4 (CNTN4) is mainly expressed in human brain. J Hum Genet. 2002;47(9):497–499. doi:10.1007/s100380200073
13. Xie T, Deng L, Mei P, et al. A genome-wide association study combining pathway analysis for typical sporadic amyotrophic lateral sclerosis in Chinese Han populations. Neurobiol Aging. 2014;35(7):1778e9–1778e23. doi:10.1016/j.neurobiolaging.2014.01.014
14. Van Dijk KR, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59(1):431–438. doi:10.1016/j.neuroimage.2011.07.044
15. Baroncini M, Jissendi P, Balland E, et al. MRI atlas of the human hypothalamus. Neuroimage. 2012;59(1):168–180. doi:10.1016/j.neuroimage.2011.07.013
16. Zhang S, Wang W, Zhornitsky S, Li CR. Resting state functional connectivity of the lateral and medial hypothalamus in cocaine dependence: an exploratory study. Front Psychiatry. 2018;9:344. doi:10.3389/fpsyt.2018.00344
17. Desport JC, Preux PM, Truong TC, et al. Nutritional status is a prognostic factor for survival in ALS patients. Neurology. 1999;53(5):1059–1063. doi:10.1212/wnl.53.5.1059
18. Desport JC, Preux PM, Magy L, et al. Factors correlated with hypermetabolism in patients with amyotrophic lateral sclerosis. Am J Clin Nutr. 2001;74(3):328–334. doi:10.1093/ajcn/74.3.328
19. Boraxbekk CJ, Stomby A, Ryberg M, et al. Diet-induced weight loss alters functional brain responses during an episodic memory task. Obes Facts. 2015;8(4):261–272. doi:10.1159/000437157
20. Yao L, Li W, Dai Z, Dong C. Eating behavior associated with gray matter volume alternations: a voxel based morphometry study. Appetite. 2016;96:572–579. doi:10.1016/j.appet.2015.10.017
21. Agosta F, Canu E, Valsasina P, et al. Divergent brain network connectivity in amyotrophic lateral sclerosis. Neurobiol Aging. 2013;34(2):419–427. doi:10.1016/j.neurobiolaging.2012.04.015
22. Verstraete E, Veldink JH, Mandl RC, van den Berg LH, van den Heuvel MP. Impaired structural motor connectome in amyotrophic lateral sclerosis. PLoS One. 2011;6(9):e24239. doi:10.1371/journal.pone.0024239
23. Tedeschi G, Trojsi F, Tessitore A, et al. Interaction between aging and neurodegeneration in amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33(5):886–898. doi:10.1016/j.neurobiolaging.2010.07.011
24. Heimrath J, Gorges M, Kassubek J, et al. Additional resources and the default mode network: evidence of increased connectivity and decreased white matter integrity in amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15(7–8):537–545. doi:10.3109/21678421.2014.911914
25. Chenji S, Jha S, Lee D, et al. Investigating default mode and sensorimotor network connectivity in amyotrophic lateral sclerosis. PLoS One. 2016;11(6):e0157443. doi:10.1371/journal.pone.0157443
26. Consonni M, Cappa SF, Dalla Bella E, Contarino VE, Lauria G. Cortical correlates of behavioural change in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2019;90(4):380–386. doi:10.1136/jnnp-2018-318619
27. Hashikawa Y, Hashikawa K, Falkner AL, Lin D. Ventromedial hypothalamus and the generation of aggression. Front Syst Neurosci. 2017;11:94. doi:10.3389/fnsys.2017.00094
28. Canteras NS. The medial hypothalamic defensive system: hodological organization and functional implications. Pharmacol Biochem Behav. 2002;71(3):481–491. doi:10.1016/s0091-3057(01)00685-2
29. Ciriello J, Caverson MM. Hypothalamic orexin-A (hypocretin-1) neuronal projections to the vestibular complex and cerebellum in the rat. Brain Res. 2014;1579:20–34. doi:10.1016/j.brainres.2014.07.008
30. Tigges J, Walker LC, Tigges M. Subcortical projections to the occipital and parietal lobes of the chimpanzee brain. J Comp Neurol. 1983;220(1):106–115. doi:10.1002/cne.902200110
31. Kamali A, Ghazi Sherbaf F, Rahmani F, et al. A direct visuosensory cortical connectivity of the human limbic system. Dissecting the trajectory of the parieto-occipito-hypothalamic tract in the human brain using diffusion weighted tractography. Neurosci Lett. 2020;728:134955. doi:10.1016/j.neulet.2020.134955
32. Zhang SQ, Fleischer J, Al-Kateb H, et al. Intragenic CNTN4 copy number variants associated with a spectrum of neurobehavioral phenotypes. Eur J Med Genet. 2020;63(3):103736. doi:10.1016/j.ejmg.2019.103736
33. Cottrell CE, Bir N, Varga E, et al. Contactin 4 as an autism susceptibility locus. Autism Res. 2011;4(3):189–199. doi:10.1002/aur.184
34. Yoshihara Y, Kawasaki M, Tamada A, et al. Overlapping and differential expression of BIG-2, BIG-1, TAG-1, and F3: four members of an axon-associated cell adhesion molecule subgroup of the immunoglobulin superfamily. J Neurobiol. 1995;28(1):51–69. doi:10.1002/neu.480280106
35. Heise C, Preuss JM, Schroeder JC, et al. Heterogeneity of cell surface glutamate and GABA receptor expression in shank and CNTN4 autism mouse models. Front Mol Neurosci. 2018;11:212. doi:10.3389/fnmol.2018.00212
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
