Back to Journals » Infection and Drug Resistance » Volume 15
Effect of Ruta graveolens Extract on the Major Virulence Factors in Methicillin Resistant Staphylococcus aureus
Authors Rezk S , Alqabbasi O, Ramadan A , Turkey M
Received 19 October 2022
Accepted for publication 23 November 2022
Published 6 December 2022 Volume 2022:15 Pages 7147—7156
DOI https://doi.org/10.2147/IDR.S393912
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Shahinda Rezk,1 Omar Alqabbasi,2 Asmaa Ramadan,3,4 Mohamed Turkey5
1Microbiology Department, Medical Research Institute, Alexandria University, Alexandria, Egypt; 2Biology Department, Faculty of Science, Benghazi University, Benghazi, Libya; 3Microbiology and Biotechnology Department, College of Pharmacy, Arab Academy for Science, Technology and Maritime Transport, Alexandria, Egypt; 4Pharmacy Department, Ministry of Health and Population, Alexandria, Egypt; 5Microbiology and Immunology Department, Faculty of Pharmacy, October 6 University, Sixth of October City, Giza, 12585, Egypt
Correspondence: Shahinda Rezk, Microbiology Department, Medical Research Institute, Alexandria University, 169 Horreya Road, Al Ibrahimeyah Qebli WA Al Hadrah Bahri, Bab Sharqi, Alexandria, 21561, Egypt, Tel +20 1023020030, Fax +20 34283543, Email [email protected]
Purpose: Rising Antibiotic Resistance has put the world in real threat. Methicillin resistant Staphylococcus aureus (MRSA), is a predominant cause of suppurative chronic skin and soft-tissue infections. Novel insights have focused the light on plant extracts. In this study, Ruta graveolens ethanolic active extract was tested for its potential anti-virulence activities in MRSA.
Materials and Methods: A total of 100 MRSA strains causing skin and soft tissue infections were isolated and antibiotic susceptibility testing was done. Ability to form biofilm was tested phenotypically. Furthermore, the antimicrobial activity of Ruta graveolens was evaluated followed by detection of its Minimum inhibitory concentration (MIC). The inhibitory activity of this extract on biofilm formation was investigated. Afterwards, we investigated its effect on the transcription of biofilm-related genes and mecA gene.
Results: All tested isolates were sensitive to Vancomycin and Linezolid while high resistance was noted with both Fusidic acid (83%) and Gentamicin (68%). (83%) of the isolates were biofilm producers. Ruta graveolens extract showed strong antimicrobial activity against the MRSA strains with MIC 0.78 mg/mL. At subinhibitory concentration (1/2 MIC), the extract had high biofilm inhibitory effects with mean inhibition (70%). Moreover, transcriptional analysis results showed that the mean percentages of inhibition in expression of mecA, icaA and icaD genes were 52.3%, 34.8% and 33.7%, respectively, in which all showed statistically significant difference (p ≤ 0.05).
Conclusion: The current study proposes the ability of Ruta graveolens extract to reduce the biofilm formation and antibiotic resistance of MRSA through downregulation of some biofilm forming genes and mecA gene which confers resistance to B-lactam antibiotics. This may decrease our reliance on antibiotics and improve our ability to effectively treat biofilm-related skin and soft-tissue infections caused by MRSA.
Keywords: ethanolic plant extract, mecA expression, biofilm formation, skin infections
Plain Language Summary
Resistance of the bacteria to many marketed antibiotics has been rising dramatically due to antibiotic abuse in the communities and the hospitals. Thus, there is a continuous urge to find new treatment options for such infections. By looking back to traditional medicine, we found that herbs can also be used to treat some infections. This study focused on the use of a herbal extract named Ruta graveolens to test its activity on a highly antibiotic resistant bacteria named MRSA. This bacteria in known to cause various infections ranging from mild skin infections to life-threatening infections. The extract was found to have a potent killing effect on MRSA and also inhibit some major virulence factors that help the bacteria to establish the infection and become antibiotic resistant.
Introduction
Staphylococcus aureus (S. aureus) is a gram-positive commensal and opportunistic bacterium colonizing different body parts, such as the nostrils, axilla, and inguinal areas. It is the causative agent of many infections in hospitals and communities, ranging from skin and soft tissue infections to potentially life-threatening bloodstream infections.1 Prevalence rates of MRSA among hospitalized patients in Egypt vary according to the geographical region, ranging from 24.4%2 to 75%.3 These prevalence rates are alarming, necessitating robust actions to combat this persistent health-care obstacle challenge.
The virulence of S. aureus has risen with existence of Methicillin resistant S. aureus (MRSA) superbug. These strains carry the mecA gene that make them resistant to almost all beta-lactam antibiotics except Ceftaroline.4 The mecA gene encodes PBP2a protein, a modified PBP with low affinity for β-lactam antibiotics.5 MRSA is a multidrug-resistant organism. It is always resistant to various antimicrobial agents, including Fusidic acid, lincosamides, tetracyclines and aminoglycosides.6
S. aureus develops biofilms on many medical devices like implants, surgical instruments, and catheters, and inside the infected host tissues as well,7 causing life-threatening illnesses. They have incredibly challenging clinical implications for health-care professionals to deal with.8
Biofilms are crucial in the progression of chronic ulcers, which is a global and widespread condition. Colonizing bacteria are inevitable, but when a wound has a tendency to persist, there is a greater likelihood that the bacteria may assemble into the more complex biofilm structures. Due to extended persistence of the inflammatory processes in addition to the physical barrier function, this may diminish the chances of wound healing.9 There are 12 distinct genes that primarily regulate biofilm formation in S. aureus; the intercellular adhesion genes are the most important (ica A, B, C, and D).10
Due to the persistent, critical, and troublesome S. aureus biofilm-associated infections, researchers needed to study and adopt proper anti-biofilm strategies. These strategies work either by inhibiting biofilm initiation or dispersing already established mature biofilms.8 Different anti-biofilm agents have been reported to inhibit and eradicate Staphylococcal biofilms. These include some antibiotics (eg, Rifampicin, Vancomycin, Azithromycin, Clindamycin),11–13 amino acids,14 anti-biofilm molecules (eg, imidazoles, indoles, carbazoles, pyrroles),15 enzymes (eg, DNAse and exopolysaccharide-degrading-enzymes),12 nanoparticles of heavy metals,16 as well as various essential oils and plant extracts.17,18 Due to their availability and safety, the development and generation of novel anti-infective MRSA agents heavily rely on natural products.19
The antimicrobial activity of natural products against different pathogens has been previously described.20 Antimicrobial mechanisms include cell membrane disruption, intracellular biomolecule damage, and biofilm inhibition. The oxygen-radical scavenging property of natural products adds merit to their usage, leading to decreased toxicity and side effects in comparison to chemical antimicrobial agents. Moreover, natural products are chemically diverse, biochemically precise, and more economic.21 However, large-scale plant extract implementation as antibacterial agents is limited by several factors. The most important factor is the poor economic investment in this field by big pharmaceutical companies, which means that such studies are carried out by research institutions, which may have limited facilities.22
This study aimed to investigate the impact of the Ruta graveolens active plant extract as a potential inhibitor of MRSA biofilm formation. In addition, transcriptional analysis of biofilm formation genes and mecA gene, responsible for B-lactams resistance, was performed before and after treatment with the plant ethanolic extract.
Materials and Methods
The study was conducted in accordance with World Medical Association Helsinki Declaration for studies on human subjects. The study then took the approval of the Ethics Committee of the Medical Research Institute, Alexandria University (IORG#: IORG0008812) and an informed written consent was obtained from the patients.
Bacterial Isolates
One hundred MRSA strains causing skin and soft tissue infections were isolated from clinical specimens from January 2022 to June 2022 that were admitted to Microbiology laboratory, Medical Research Institute hospital. These samples were isolated as a part of the routine hospital laboratory procedure. MRSA strains were isolated by culturing on Blood agar, Staphylococcus Chromogenic Agar, and MRSA Chromogenic Agar (Condalab, Madrid, Spain). Primary identification was performed by colony morphology and then microscopically by Gram-stained smears. Further identification and antibiotic susceptibility testing were done by Vitek 2 Compact system (bioMérieux, France) according to the manufacturer’s protocol. Aliquots of each isolate were inoculated in LB-Glycerol and stored in the freezer at −80 C for later investigations.
Phenotypic Detection of Biofilm Formation
Quantitative detection of biofilm formation was performed based on the microtiter plate method of Stepanović et al.23 Overnight bacterial cultures were grown in Tryptone soya broth (TSB). Cultures were calibrated to 0.5 McFarland turbidity standard and diluted 1:100 with fresh TSB. Three wells of a sterile 96-well, flat-bottomed plastic plate with a lid were inoculated with 200 μL of bacterial suspension each. The plates statically incubated at 37°C for 24 hours in aerobic conditions (negative control wells were included - containing broth only). S. aureus ATCC 25923 standard strain was used as a positive control. After incubation, the content of each well was aspirated, and each well was gently washed with 250 μL of sterile phosphate-buffered saline three times; all non-adherent bacteria were removed by shaking the plates. Fixation of the remaining attached bacteria was established by the addition of 200 μL of 99% methanol per well. After 15 min fixation, the plates were emptied and left to dry. Wells were then stained with 200 μL of 2% Hucker crystal violet for 15 minutes. Excess stain was rinsed off by soaking the plate in distilled water; plates were removed and left to air dry. The dye bound to the adherent cells was resolubilized with 160 μL of 33% (v/v) glacial acetic acid per well and left for 10–15 min. Finally, the optical density (OD) of was measured at 570 nm using an automated ELISA reader (TECAN, Switzerland); 33% glacial acetic acid was used as blank (negative control). Each growing strain was tested in triplicates, and the average reading of the three wells was obtained.
Adherence capabilities of tested strains were assessed. Strains were classified into 4 categories: non-adherent (OD ≤ ODc), weakly adherent (ODc < OD ≤ 2×ODc), moderately adherent (2×ODc < OD ≤ 4×ODc), or strongly adherent (OD > 4×ODc), based upon the calculated cut-off OD (ODc) values of the bacterial films. The cut-off OD (ODc) for the microtiter-plate test is defined as three standard deviations above the mean OD of the negative control.
Phytochemical Studies
Herbal Extract Preparation and Characterization
Ruta graveolens plant (Family Rutaceae) was obtained from an Experimental Station of Aromatic and Medicinal Plants, Faculty of Pharmacy, University of Cairo, Egypt during October 2021. Dr. AbdelHalem Abdelmogali; an expert taxonomist at research centre for agriculture (Giza, Egypt) has kindly verified the taxonomical plant features. The voucher specimens were deposited in the herbarium of Pharmacognosy Department, Faculty of Pharmacy, October 6 University, and given the code (Rg-01). The plant’s aerial part (leaves and flowers) was first air-dried for two weeks and coarsely powdered. About 200–300 g of the dried powder was extracted using 70% ethanol by maceration; the process was repeated several times until total plant exhaustion. A rotary-evaporator was subsequently used to concentrate and filter the ethanolic extract solution (while operating under decreased pressure and at a temperature below 50°C). After being dried to leave a solid residue, the extract was kept at −20°C in a tightly sealed glass container until it was tested for its antibacterial activity.
Screening for the Antimicrobial Activity of Ruta graveolens
Screening for the bactericidal activity of the natural plant extract was carried out using the cup-plate method.24 Fresh cultures of MRSA isolates, calibrated at a cell density of approximately 0.5 McFarland, were streaked onto Muller Hinton agar plates. Cups (7 mm) were made aseptically using a sterile cork borer. The previously prepared plant extract was dissolved in 0.5% dimethyl sulfoxide (DMSO) to get a 100 mg/mL solution. Accurately, 200 µL was pipetted into cups made in these plates. In order to neutralize the antimicrobial effects of DMSO, wells containing only DMSO were utilized as a negative control. After 24 hours incubation of the inoculated plates at 37°C, the diameters of inhibition zones were recorded, reflecting the activity of the extract.
Detection of Minimum Inhibitory Concentration (MIC) Using Broth Microdilution Method
MIC of the Ruta graveolens active extract was tested by broth microdilution method, using three strong-biofilm forming antibiotic-resistant MRSA strains. Flat-bottomed sterile 96-well microtiter plates were used to perform the experiments. Tests were performed using Muller Hinton Broth (MHB) medium. The plant extract was previously prepared and adjusted as 100 mg/mL in dimethyl sulfoxide (DMSO), then two-fold serial dilutions of the plant extract were made, reaching final concentrations ranging from 50–0.049 mg/mL. One hundred µL of overnight bacterial cultures of each strain, calibrated to approximately 0.5 McFarland was inoculated onto 100 µL of the plant extract dilutions, and the plate was then incubated for 24 hours at 37 C. The lowest concentration of the plant extract at which the microorganism does not demonstrate visible growth is defined as the MIC. For each concentration, the extract was tested in triplicate, and the experiment was done 3 times separately. According to the CLSI 2022, MIC was evaluated using the broth microdilution method.25,26
Effect of a Sub-Inhibitory Concentration of Ruta graveolens on Biofilm-Formation
Using the previously described microtiter plate technique, the anti-biofilm activity of the extract against the selected isolates was assessed at the sub-MIC extract concentration (1/2 MIC); extract-free wells served as controls. The following formula was used to quantify the percentage of biofilm inhibition:
Transcriptional Analysis
Pure bacterial colonies from freshly-streaked plates of the selected MRSA isolates grown on Luria–Bertani (LB) agar were inoculated in liquid LB broth, in the presence and absence of the sub-MIC of the extract. Cultures were grown in a shaking incubator at 225 rpm/37℃. After overnight incubation, 10 mL of each inoculum was transferred to a sterile falcon tube and centrifuged in a cooling centrifuge (4℃) for 5 min at 5000 rpm. The supernatants were discarded, and the pellets were kept at −80 ℃ for later RNA extraction.
RNA Extraction and Reverse Transcription
Ready-to-use Invitrogen™ TRIzol™ Reagent (15596026, Life Technologies, USA) was utilized for total-RNA purification, according to the manufacturer’s protocol.27 QuantiTects Reverse Transcription Kit (Qiagen, USA) was used to reverse transcribe 1 μg of total RNA using a random primer hexamer in a two-step reaction. Any genomic DNA (gDNA) contamination was excluded using a Wipeout buffer.
Real-Time PCR Assay for Gene Expression Analysis
Specific primer pairs were used to amplify total cDNA (30 ng) template (Table 1), using Maxima SYBR Green/Fluorescin qPCR Master Mix (2X, Thermo Scientific, Waltham, MA, USA). The thermal profile was: initial denaturation at 95°C for 5 minutes, then 35 cycles at 94°C for 30 seconds, 55°C for 30 seconds, 72°C for 30 seconds, and final extension step for 5 minutes at 72°C. Samples were subjected to quantitative real-time PCR in duplicates, and then the mean values were used for subsequent analysis. Rotor-Gene Q MDx (Qiagen, USA) performed the amplification, automatically collected the data, and afterwards analyzed the value of threshold Cycle (Ct), which was normalized to an average Ct value of the housekeeping gene (∆Ct). The relative gene expression fold change was estimated using 2−∆∆ct method,28 standardized to the reference housekeeping gene 16S rRNA.
 |
Table 1 List of Primer Sequences Used for Quantitative Real-Time PCR (qRT-PCR) |
Statistical Analysis
The statistically significant relationship between the untreated control and the tested isolates was done by a one-way variance A test. P ≤ 0.05 demonstrated a significant difference.
Results
Bacterial Isolates
One hundred MRSA strains were isolated from clinical specimens identified phenotypically and automatically using Vitek 2 Compact System. A total of 50 (50%) isolates were obtained from aspirated pus, 28 (28%) from surgical chronic wounds, and 22 (22%) from superficial abscesses.
Antimicrobial Susceptibility Testing (AST)
AST and Cefoxitin screens were performed using Vitek 2 Compact System. This study revealed that all isolates were positive for the Cefoxitin screen test, thus interpreted as MRSA. All tested isolates were sensitive to Vancomycin and Teicoplanin, with minimum inhibitory concentration (MIC) ≤0.5 mg/L for each of them, and also to Linezolid with MIC 2 mg/L. High resistance was noted with both Fusidic acid (83%) and Gentamicin (68%) with MIC 8 and ≥16 mg/L, respectively (Table 2). Inducible Clindamycin resistance was detected in 5% of the tested isolates.
 |
Table 2 Antimicrobial Susceptibility of the Isolates Against Corresponding Antibiotics |
Biofilm Formation
This study revealed that 83% of the MRSA isolates were biofilm producers while 17% were non-biofilm formers. It also distinguished among strong, moderate, and weak biofilm-producing strains, as shown in Table 3.
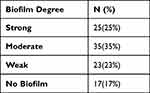 |
Table 3 The Degree of Biofilm Formation Among the MRSA Isolates |
Antimicrobial Activity of Ruta graveolens Extract on MRSA Isolates
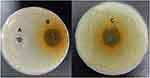
Inhibition zones ranging from 26 to 30 mm were recorded around preformed wells in the Muller Hinton agar plates streaked with 0.5 McFarland of the MRSA isolates and incubated for 24 hours at 37 ͦC, as shown in Figure 1. Each well contained 200 ul of dissolved Ruta graveolens extract. Small inhibition zones were detected with the negative wells containing only DMSO, ranging from 10 to 11 mm.
Determination of MIC of the Ruta graveolens Extract on MRSA Isolates
The MIC of the dissolved plant extract was tested against three selected MRSA isolates, which were strong biofilm producers and had high antibiotic resistance, named Test 1, Test 2, and Test 3. The three isolates showed the same MIC, 0.78 mg/mL.
Effect of Sub-Inhibitory Concentration of the Extract on Biofilm-Formation
The sub-MIC (1/2 MIC) of Ruta graveolens extract showed inhibition on the biofilm-formation of the three selected clinical isolates by 70.1%, 77.7%, and 62.2% in Test 1, Test 2, and Test 3, respectively, compared to the untreated control. The three treated isolates displayed significant inhibition in biofilm production.
Effect of Ruta graveolens Extract on mecA, icaA and icaD Genes Transcription Levels
mecA Gene
The percent of inhibition of mecA gene in the three selected treated MRSA isolates were 50%, 53%, and 54% in Test 1, Test 2, and Test 3, respectively, normalized to the housekeeping gene 16S rRNA (Figure 2). There was a statistically significant difference between the untreated control and the treated isolates (p ≤ 0.05).
 |
Figure 2 Effect of Ruta graveolens extract on the transcriptional level of mecA gene in untreated control and treated MRSA isolates. |
icaA Gene
The percent of inhibition of icaA gene in the same MRSA isolates were 33.6%, 34.5%, and 36.3%, in Test 1, Test 2, and Test 3, respectively, normalized to the housekeeping gene 16S rRNA (Figure 3). There was a statistically significant difference between the untreated control and the treated isolates (p ≤ 0.05).
 |
Figure 3 Effect of Ruta graveolens extract on the transcriptional level of icaA gene in untreated control and treated MRSA isolates. |
icaD Gene
The influence of the herbal extract on icaD gene was also evaluated. The percent of inhibition of icaD gene were 33.33%, 35.5%, and 32.2%, in Test 1, Test 2, and Test 3, respectively, normalized to the housekeeping gene 16S rRNA (Figure 4). There was a statistically significant difference between the untreated control and the treated isolates in the relative (p ≤ 0.05).
 |
Figure 4 Effect of Ruta graveolens extract on the transcriptional level of icaD gene in untreated control and treated MRSA isolates. |
Discussion
In developing nations, it is becoming routine practice to abuse and overprescribe antibiotics. The availability of over-the-counter antibiotics and unregulated sales of antibiotics in underdeveloped nations have all contributed to an exponential rise in drug resistance. The desperate need for new compounds with potential antibacterial activities against various drug-resistant microbial strains has been driven by the alarming rise in antimicrobial drug resistance.29 One of the most prevalent multidrug-resistant organisms is MRSA. It always causes skin infections in the community. If left untreated, infections can cause sepsis. Due to the rising resistance levels to topical fusidic acid, which was preferably used to treat MRSA skin infections, we have directed our attention to testing some plant extracts that can be potentially used as a part of a topical treatment. The belief that some plants have medicinal properties has a long history and was widespread throughout the majority of ancient cultures. It is a part of the national and social traditions of various societies.30
Ancient societies were familiar with the medicinal herb Ruta graveolens. Asian and European scientists utilize it to treat a wide range of illnesses, including seizures, cough, hypertension and wound healing. Numerous investigations have noted the antimicrobial activity of this plant’s extracts on bacteria, fungi, protozoa, worms, and protozoa, although the mechanisms are not fully understood.31 Therefore, this study aimed to investigate the antibacterial effects of medicinal extracts of Ruta graveolens on clinical MRSA isolates.
Elevated resistance was observed with both Fusidic acid (83%) and Gentamicin (68%) among the 100 clinical MRSA isolates. Several studies have also reported high resistance levels for these two antibiotics.32–34 Consequently, these isolates show resistance to most topical antibiotics in the market targeting Gram-positive bacteria.
Numerous studies have reported the high prevalence of biofilm formation among MRSA isolates.35–37 The development of biofilm at the site of a wound is crucial to its chronicity. Due to the difficulties of eliminating it, scientists have focused their efforts on developing novel compounds that can interact with biofilm to speed up the healing process.9 Taking into account MRSA antibiotic resistance, virulence, propensity to tenaciously colonize wounds by biofilms, and rising infections in both hospital and community settings, this study focused on R. graveolens ethanolic extract, which could have an anti-MRSA effect.
Many previous studies had described the antibacterial activity of this plant extract on Staphylococcus aureus,26,38,39 but to the best of our knowledge, it has not yet been associated directly with any specific anti-biofilm formation and mecA expression downregulation.
This study had showed the inhibitory effect of the sub-MIC of R. graveolens extract on biofilm formation capacity of strong biofilm-former MRSA strains compared to the untreated control. Furthermore, to verify its effect on gene expression of the biofilm-forming genes icaA and icaD, the expression levels were assessed after treatment with the sub-MIC (1/2 MIC) as well. There was a statistically significant difference between the untreated control and the treated isolates (p≤0.05) regarding the inhibition percentages in the expression of both genes. The phenotypic inhibition of the biofilm formation was more evident than the genotypic inhibition in the three isolates. This can be explained by the presence of many other biofilm formation genes that the drug may have acted upon. Thus, further studies on other biofilm formation genes will be of utmost importance to pave the way for new medicinal treatment options.
Additionally, a statistically significant difference in the percentage of inhibition in mecA gene expression was documented between treated isolates and the untreated control (p≤ 0.005).
Accordingly, based on the phenotypic impact on biofilm formation and the genotypic findings, R. graveolens can be claimed to have strong inhibitory effects on virulence factors of MRSA.
Conclusion
These results suggest that R. graveolens ethanolic extract could be developed as a promising anti-MRSA agent by affecting biofilm formation and acting on the mecA gene, which limits its action against B-lactams. It might be incorporated into topical antibacterial agents used in treating wounds infected with MRSA as a part of a combined treatment regimen to decrease the chances of resistance development and to get over the infection so as to promote wound healing.
Acknowledgment
The authors thank Dr. AbdelHalem Abdelmogali who had confirmed the taxonomical features of the plant extract at the agriculture research center, Giza, Egypt.
Disclosure
The authors report no conflicts of interest in relation to this work. No organizations funded this research and there are no financial interests.
References
1. Oliveira D, Borges A, Simões M. Staphylococcus aureus toxins and their molecular activity in infectious diseases. Toxins. 2018;10(6):252. doi:10.3390/toxins10060252
2. El-Gendy MMAA, El-Bondkly AMA, Keera AA, Ali AM. Incidence of methicillin-resistant Staphylococcus aureus (MRSA) in microbial community of cancer patients and evaluation of their resistant pattern. Arab J Sci Eng. 2018;43(1):83–92. doi:10.1007/s13369-017-2670-4
3. Algammal AM, Hetta HF, Elkelish A, et al. Methicillin-resistant Staphylococcus aureus (MRSA): one health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect Drug Resist. 2020;13:3255. doi:10.2147/IDR.S272733
4. Cosimi RA, Beik N, Kubiak DW, Johnson JA. Ceftaroline for severe methicillin-resistant Staphylococcus aureus infections: a systematic review. Open Forum Infect Dis. 2017;4(2):ofx084. doi:10.1093/ofid/ofx084
5. Gnanamani A, Hariharan P, Paul- Satyaseela M. Staphylococcus aureus: overview of bacteriology, clinical diseases, epidemiology, antibiotic resistance and therapeutic approach. In: Enany S, Alexander L, editors. Frontiers in Staphylococcus Aureus. London: IntechOpen; 2017:3–26.
6. Gajdács M, Urbán E. Epidemiology and resistance trends of Staphylococcus aureus isolated from vaginal samples: a 10-year retrospective study in Hungary. Acta Dermatovenerol Alp Panon Adriat. 2019;28(4):143–147. doi:10.15570/actaapa.2019.35
7. Khatoon Z, McTiernan CD, Suuronen EJ, Mah TF, Alarcon EI. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon. 2018;4(12):e01067. doi:10.1016/j.heliyon.2018.e01067
8. Idrees M, Sawant S, Karodia N, Rahman A. Staphylococcus aureus biofilm: morphology, genetics, pathogenesis and treatment strategies. Int J Environ Res Public Health. 2021;18(14):7602. doi:10.3390/ijerph18147602
9. Simonetti O, Marasca S, Candelora M, et al. Methicillin-resistant Staphylococcus aureus as a cause of chronic wound infections: alternative strategies for management. AIMS Microbiol. 2022;8(2):125–137. doi:10.3934/microbiol.2022011
10. Nourbakhsh F, Namvar AE. Detection of genes involved in biofilm formation in Staphylococcus aureus isolates. GMS Hyg Infect Control. 2016;11:Doc07. doi:10.3205/dgkh000267
11. Hu H, Ramezanpour M, Hayes AJ, et al. Sub-inhibitory clindamycin and azithromycin reduce S. aureus exoprotein induced toxicity, inflammation, barrier disruption and invasion. J Clin Med. 2019;8(10):1617. doi:10.3390/jcm8101617
12. Sharma D, Misba L, Khan AU. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control. 2019;8(1):76. doi:10.1186/s13756-019-0533-3
13. Wells CM, Beenken KE, Smeltzer MS, Courtney HS, Jennings JA, Haggard WO. Ciprofloxacin and rifampin dual antibiotic-loaded biopolymer chitosan sponge for bacterial inhibition. Mil Med. 2018;183(suppl_1):433–444. doi:10.1093/milmed/usx150
14. Hochbaum AI, Kolodkin-Gal I, Foulston L, Kolter R, Aizenberg J, Losick R. Inhibitory effects of D-amino acids on Staphylococcus aureus biofilm development. J Bacteriol. 2011;193(20):5616–5622. doi:10.1128/JB.05534-11
15. Parrino B, Schillaci D, Carnevale I, et al. Synthetic small molecules as anti-biofilm agents in the struggle against antibiotic resistance. Eur J Med Chem. 2019;161:154–178. doi:10.1016/j.ejmech.2018.10.036
16. Fontecha-Umaña F, Ríos-Castillo AG, Ripolles-Avila C, Rodríguez-Jerez JJ. Antimicrobial activity and prevention of bacterial biofilm formation of silver and zinc oxide nanoparticle-containing polyester surfaces at various concentrations for use. Foods. 2020;9(4):442. doi:10.3390/foods9040442
17. Bazargani MM, Rohloff J. Antibiofilm activity of essential oils and plant extracts against Staphylococcus aureus and Escherichia coli biofilms. Food Control. 2016;61:156–164. doi:10.1016/j.foodcont.2015.09.036
18. Monteiro-Neto V, de Souza CD, Gonzaga LF, et al. Cuminaldehyde potentiates the antimicrobial actions of ciprofloxacin against Staphylococcus aureus and Escherichia coli. PLoS One. 2020;15(5):e0232987. doi:10.1371/journal.pone.0232987
19. Craft KM, Nguyen JM, Berg LJ, Townsend SD. Methicillin-resistant Staphylococcus aureus (MRSA): antibiotic-resistance and the biofilm phenotype. Med Chem Comm. 2019;10(8):1231–1241. doi:10.1039/C9MD00044E
20. Nogueira JWA, Costa RA, da Cunha MT, Cavalcante TTA. Antibiofilm activity of natural substances derived from plants. Afr J Microbiol Res. 2017;11(26):1051–1060. doi:10.5897/AJMR2016.8180
21. Parham S, Kharazi AZ, Bakhsheshi-Rad HR, et al. Antioxidant, antimicrobial and antiviral properties of herbal materials. Antioxidants. 2020;9(12):1309. doi:10.3390/antiox9121309
22. Cushnie T, Cushnie B, Echeverría J, et al. Bioprospecting for antibacterial drugs: a multidisciplinary perspective on natural product source material, bioassay selection and avoidable pitfalls. Pharm Res. 2020;37(7):1–24. doi:10.1007/s11095-020-02849-1
23. Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40(2):175–179. doi:10.1016/S0167-7012(00)00122-6
24. Barry A. The Antimicrobic Susceptibility Test: Principles and Practices. Philadelphia: Lippincott Williams & Wilkins; 1976.
25. Clinical and Laboratory Standards Institute (CLSI). M100: Performance Standards for Antimicrobial Susceptibility Testing.
26. Ojala T, Remes S, Haansuu P, et al. Antimicrobial activity of some coumarin containing herbal plants growing in Finland. J Ethnopharmacol. 2000;73(1–2):299–305. doi:10.1016/S0378-8741(00)00279-8
27. Simms D, Cizdziel P, Chomczynski P. TRIzol: a new reagent for optimal single-step isolation of RNA. Focus. 1993;15(4):532–535.
28. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
29. Al-Majmaie S, Nahar L, Rahman M, et al. Anti-MRSA constituents from Ruta chalepensis (Rutaceae) grown in Iraq, and in silico studies on two of most active compounds, chalepensin and 6-hydroxy-rutin 3’,7-dimethyl ether. Molecules. 2021;26(4):1114. doi:10.3390/molecules26041114
30. Honarmand H, Saeidinia A. Study on antibacterial effect of Ruta graveolens extracts on pathogenic bacteria. Ann Biol Res. 2012;3:4542–4545.
31. Naghibi Harat Z, Kamalinejad M, Sadeghi M, Sadeghipour E. A review on Ruta graveolens L. its usage in traditional medicine and modern research data. J Med Plants. 2009;8:1–19.
32. Hajikhani B, Goudarzi M, Kakavandi S, et al. The global prevalence of fusidic acid resistance in clinical isolates of Staphylococcus aureus: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2021;10(1):75. doi:10.1186/s13756-021-00943-6
33. Wang JT, Huang IW, Chang SC, et al. Increasing resistance to fusidic acid among clinical isolates of MRSA. J Antimicrob Chemother. 2017;72(2):616–618. doi:10.1093/jac/dkw430
34. Rahimi F. Characterization of resistance to aminoglycosides in methicillin-resistant Staphylococcus aureus strains isolated from a tertiary care hospital in Tehran, Iran. Jundishapur J Microbiol. 2016;9(1):e29237. doi:10.5812/jjm.29237
35. Silva V, Almeida L, Gaio V, et al. Biofilm formation of multidrug-resistant MRSA strains isolated from different types of human infections. Pathogens. 2021;10(8):970. doi:10.3390/pathogens10080970
36. Leshem T, Schnall BS, Azrad M, Baum M, Rokney A, Peretz A. Incidence of biofilm formation among MRSA and MSSA clinical isolates from hospitalized patients in Israel. J Appl Microbiol. 2022;133(2):922–929. doi:10.1111/jam.15612
37. Cascioferro S, Carbone D, Parrino B, et al. Therapeutic strategies to counteract antibiotic resistance in MRSA biofilm-associated infections. Chem Med Chem. 2021;16(1):65–80. doi:10.1002/cmdc.202000677
38. Ivanova A, Mikhova B, Najdenski H, Tsvetkova I, Kostova I. Antimicrobial and cytotoxic activity of Ruta graveolens. Fitoterapia. 2005;76(3–4):344–347. doi:10.1016/j.fitote.2005.02.008
39. Alzoreky NS, Nakahara K. Antibacterial activity of extracts from some edible plants commonly consumed in Asia. Int J Food Microbiol. 2003;80(3):223–230. doi:10.1016/S0168-1605(02)00169-1
40. Duran N, Ozer B, Duran GG, Onlen Y, Demir C. Antibiotic resistance genes and susceptibility patterns in staphylococci. Indian J Med Res. 2012;135(3):389–396.
41. Arciola CR, Baldassarri L, Montanaro L. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J Clin Microbiol. 2001;39(6):2151–2156. doi:10.1128/JCM.39.6.2151-2156.2001
42. Inoue M, Suzuki T, Fujita Y, et al. Synergistic effect of polyoxometalates in combination with oxacillin against methicillin-resistant and vancomycin-resistant Staphylococcus aureus: a high initial inoculum of 1 x 108 cfu/mL for in vivo test. Biomed Pharmacother. 2006;60(5):220–226. doi:10.1016/j.biopha.2006.04.006
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.