Back to Journals » International Journal of General Medicine » Volume 15
Effect of PFKFB4 on the Prognosis and Immune Regulation of NSCLC and Its Mechanism
Authors Zhou Y , Fan Y, Qiu B, Lou M, Liu X, Yuan K, Tong J
Received 5 April 2022
Accepted for publication 25 July 2022
Published 2 August 2022 Volume 2022:15 Pages 6341—6353
DOI https://doi.org/10.2147/IJGM.S369126
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Yong Zhou,1,* Yongfei Fan,1,* Binzhe Qiu,1 Ming Lou,1 Xiaoshuang Liu,2 Kai Yuan,1,3 Jichun Tong1
1The Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University, Changzhou, People’s Republic of China; 2Nanjing Jinling Hospital: East Region Military Command General Hospital, Nanjing, People’s Republic of China; 3Heart and Lung Disease Laboratory, The Affiliated Changzhou No.2 People’s Hospital of Nanjing Medical University, Changzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jichun Tong; Kai Yuan, Email [email protected]; [email protected]
Background: NSCLC (non-small cell lung cancer) has become the malignancy with the highest incidence and mortality rate worldwide. Fructose-6-phosphate 2-kinase/fructose-2,6-bisphosphatase 4 (PFKFB4) is a key regulator of glycolysis with both kinase and phosphatase activities. The Warburg effect, or increased glycolysis in tumors, provides the metabolic basis for cancer cell proliferation and metastasis, and the Warburg pathway enzyme PFKFB4 is a newly identified important kinase. This study aimed to elucidate the poor prognostic relevance of PFKFB4 in non-small cell lung cancer tissues and its relationship with immune cell infiltration, immune cell biomarkers, and immune checkpoints.
Methods: In this study, immunohistochemical methods were used to assess PFKFB4 expression levels in 140 surgical specimens from patients with histologically confirmed non-small cell lung cancer and to investigate the relationship between PFKFB4 expression levels and the patients’ clinicopathological characteristics. The impact of PFKFB4 expression on prognosis was evaluated using Kaplan–Meier survival analysis and Cox regression analysis.
Results: When compared to normal paracrine tissues, PFKFB4 expression was enhanced in lung cancer tissues, and Kaplan–Meier survival analysis revealed that patients with high PFKFB4 expression had a worse prognosis. In NSCLC, PFKFB4 was found to be associated with immune cell infiltration and immunological checkpoints.
Conclusion: PFKFB4 expression may be upregulated as a sign of poor prognosis in NSCLC, and PFKFB4 may be implicated not only in the genesis and progression of NSCLC but also in its immunological control.
Keywords: PFKFB4, NSCLC, prognostic marker, immune cells
Introduction
Lung cancer is one of the most common malignancies, and according to the latest global epidemiological yearbook of tumors, it remains the most prevalent and deadliest malignancy worldwide, compared to five years ago. One of the reasons for this is that there are no molecular markers that are highly effective in indicating the treatment or prognosis of lung cancer.1 Although there has been some improvement in the prognosis of NSCLC patients over the past two decades with increased tobacco prevention measures and the availability of molecularly targeted drugs, improvements in outcome and survival have only occurred in a subset of patients, and for most of the new molecularly targeted drugs, there is a lack of clinically predictive markers to guide individualized treatment, optimize drug efficacy and prolong patient survival. Therefore, it is important to identify new molecular markers and therapeutic targets for lung cancer.
Cancer metabolism research has revealed fresh insights into the adaptive processes of cancer cells, namely the increased demand for energy and biosynthetic precursors that alter their metabolic activities to satisfy their own needs.2 The Warburg effect occurs when glycolysis during tumorigenesis produces significant levels of lactic and pyruvic acid, which is a typical characteristic of aggressive cancer cells.3 It is caused by increased glycolysis in the presence of oxygen, which is accompanied by an increase in the rate of glucose transport.4 Under the control of the hypoxia-inducible factor, high rates of glycolysis increase the production of glycolytic enzymes and glucose transport proteins (HIF).5 Hypoxia is one of the most effective inducers of the expression of genes involved in maintaining cellular energy,6 especially those involved in glycolysis. Warburg pathway enzyme fructose-6-phosphate 2-kinase/fructose-2,6-bisphosphatase 4 (PFKFB4) is a newly identified key factor in the regulation of transcriptional reprogramming.7 PFKFB4 has been reported to be overexpressed in gastric, breast, thyroid, bladder, and colon cancer tissues; however, relatively little is known about the function and biological role of PFKFB4 in NSCLC.
In this study, the relationship between PFKFB4, clinicopathological variables, and survival was analysed using an immunohistochemical (IHC) approach. To learn more about the role of PFKFB4 in NSCLC diagnosis and prognosis prediction, as well as its link to DNA methylation, immune cell infiltration, and immunological checkpoints.
Materials and Methods
Datasets and Data Availability
The Cancer Genome Atlas (TCGA: https://cancergenome.nih.gov) was used to extract the sex, age, survival, status, topography, and tumor-node-metastasis (TNM) stage of lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) samples. A total of 513 TCGA-LUAD and 501 TCGA-LUSC samples were qualified and utilized for further analysis. Genotype-Tissue Expression (GTEx) (https://gtexportal.org/home/datasets) also collected data from normal lung tissue samples.
Patients and Specimens
The clinical data and surgical specimens of 140 patients diagnosed with stage I–III primary non-small cell lung cancer in the Department of Thoracic Surgery, Changzhou Second People’s Hospital, were collected retrospectively from March 2003 to March 2006. The patients did not receive chemotherapy, radiotherapy, or other antitumor treatments before surgery. All patients signed informed consent prior to specimen collection, and TNM staging was evaluated according to the criteria in the International Association for the study (IAS) staging procedure. The study was approved by the Research Ethics Review Committee of Changzhou Second People’s Hospital.
Tissue Microarray and Immunohistochemical Staining
140 surgical lung cancer carcinoma specimens were fixed using a standard process and tissue microarrays (TMAs) were created. After that, formalin-fixed paraffin-embedded (FFPE) samples that had been archived and de-identified were examined. A rabbit anti-PFKFB4 polyclonal antibody [ab137785] was used to detect PFKFB4 using a two-step approach for immunohistochemical staining of TMAs for PFKFB4 (1:50; Abcam). Instead of primary antibodies, phosphate-buffered saline was used to provide negative controls. Positive controls were created using the instructions that came with the antibodies. The immunoreactivity score (IRS) of PFKFB4 was calculated using the proportion of positive cells and the staining intensity (SI) (PP), IRS=SI*PP, for example. Colorless gets 0 points, pale yellow gets 1 point, brown gets 2 points, and tan gets 3 points. PP: 0 for negative cells, 1 for positive cells with a 10% chance of being positive, 2 for 11% with a 50% chance of being positive, 3 for 51% with a 100% chance of being positive.
Correlation Analysis of PFKFB4 with Tumor Staging and Prognosis
Correlation analysis of PFKGB4 with tumor staging (T-stage, pathological stage, histological grade, AFP level) was performed using the R package and diagnostic ROC curves were established. Survival analysis was performed using the Kaplan - Meier method.
DNA Methylation Information for PFKFB4
The MethSurv database (https://biit.cs.ut.ee/methsurv/) was applied to analyze the DNA methylation sites of PFKFB4 in the TCGA database. In addition, we assessed the prognostic value of CpG methylation in PFKFB4, with survival outcome as overall survival (OS).
Correlation Analysis of PFKFB4 with Immune Cell Infiltration and Immune Checkpoints
The immune response of NSCLC patients to 22 PFKFB4, which includes 22 immune cell types, was assessed using data from CIBERSORT (https://cibersort.stanford.edu/). The TIMER database was used to look for a link between PGKFB4 expression in NSCLC and immune cell infiltration and markers, as well as a link between PGKFB4 expression in NSCLC and immunological checkpoints. Statistical significance was defined as p< 0.05.8
Statistical Analysis
SPSS version 22.0 software was used to process the data, and GraphPad Prism version 7.0 software was used to produce the images. Count data were compared using the McNamara test, chi-square test or Wilcoxon rank-sum test, survival analysis by Kaplan-Meier method in parallel with Log rank test, and multifactorial analysis by Cox proportional risk model. Differences were considered statistically significant at P < 0.05.
Results
Clinicopathological Features of Patients with NSCLC
A total of 140 NSCLC and matched adjacent normal tissues were collected to analyze the expression levels of PFKFB4 for clinical features of non-small cell lung cancer. The clinicopathological characteristics of this cohort are summarized in Tables 1 and 2. We used immunohistochemical labeling for PFKFB4 expression in tumor samples to investigate the clinical importance of PFKFB4 in lung cancer. The majority of outer stromal cells were PFKFB4 negative. Positive staining for PFKFB4 protein occurred mostly in the cytoplasm of non-small cell lung cancer cells (Figure 1A–D).
 |
Table 1 Characteristics of All Patients with NSCLC |
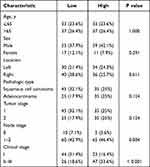 |
Table 2 Correlation Between PFKFB4 Expression and Clinicopathologic Features in Patients with Non-Small Cell Lung Cancer |
PFKFB4 Expression Levels in NSCLC
The expression of PFKFB4 in NSCLC tissues was analyzed based on TCGA data. The results showed that PFKFB4 expression was upregulated in non-small cell lung cancer tissues. PFKFB4 was expressed at higher levels in lung squamous cell carcinoma (LUSC), NSCLC, and lung adenocarcinoma (LUAD) tissues than in paired related paraneoplastic tissues; These differences were statistically significant (p< 0.05) (Figure 2A–C). PFKFB4 expression was not correlated with N phase. (Figure 2D). PFKFB4 expression was significantly correlated with the T stage (Figure 2E). PFKFB4 expression was not correlated with Clinical stage (Figure 2F). The late survival rate was lower in non-small cell lung cancer with high PFKFB4 expression.
Univariate and Multivariate Analyses for Prognosis in NSCLC Patients
Univariate analysis showed that high PFKFB4 expression and tumor grade was related to shorter outcome survival (OS) in patients with NSCLC (P<0. 05). High PFKFB4 expression and tumor grade were related to shorter OS (P<0. 05). The multifactorial results showed that PFKFB4 differential expression and lymph node metastasis were independent factors influencing the OS, as listed in Table 3.
 |
Table 3 Univariate and Multivariate Cox Regression Analyses of Various Prognostic Parameters in Patients with NSCLC |
Relationship Between PFKFB4 Expression and NSCLC Survival
The study’s follow-up began in April 2005 and continued until April 2013, with a median follow-up of 96 months. The difference in survival time between NSCLC patients with high and low PFKFB4 expression was investigated using Kaplan-Meier survival analysis. The results showed that patients with NSCLC with high PFKFB4 expression, LUSC, and LUAD had longer survival than those with low PFKFB4 expression, and the difference was highly statistically significant (p < 0.01) (Figure 2G–I).
Diagnostic Value of PFKFB4 Expression in NSCLC
We calculated the area under the curve subject to the working characteristic curves, which were 0744 (95% confidence interval: 0.714–0.774) in NSCLC (Figure 3A), 0.869 (95% confidence interval: 0.837–0.900) in LUSC (Figure 3B), and 0.692 (95% confidence interval: 0.646–0.739) in LUAD (Figure 3C). These results indicated that PFKFB4 expression had good specificity and sensitivity for the diagnosis of NSCLC, LUSC, and LUAD. We then plotted PFKFB4 as a risk score on a bar graph and added each score to get the overall score; greater total scores indicated worse prognoses (Figure 3D). The calibration plots for the 1-, 3-, and 5-year forecasts were likewise closer to the actual data, demonstrating that the PFKFB4 prognostic model was accurate in predicting the prognosis of NSCLC patients (Figure 3E).
 |
Figure 3 (A–C) ROC curves of PFKFB4 expression in NSCLC, LUAD and LUSC. (D) Line graph of NSCLC prognostic model. (E) NSCLC prognostic model 1-year and 3-year 5-year calibration curves. |
Methylation of PFKFB4 in Patients with Non-Small Cell Lung Cancer
In NSCLC patients, the MethSurv tool was utilized to investigate PFKFB4 methylation levels and the predictive value of each particular CpG. MethSurv findings revealed 33 methylated CpG sites, with the highest levels of DNA methylation found at cg21141812, cg207321140, cg25845890, and cg16902559 (Figure 4) (p<0.05). Patients with hypermethylation of PFKFB4 at these CpG sites exhibited a lower overall survival rate than those with hypermethylation of PFKFB4.
 |
Figure 4 Visualization between methylation levels and PFKFB4 expression. |
PFKFB4 Expression About Immune Cell Infiltration and Immune Checkpoints
Using the CIBERSORT algorithm to observe the clustering of 22 tumor immune cells in NSCLC tissues (Figure 5A), there were significant differences in the proportion and subpopulation distribution of the above tumor immune cells in the high and low PFKFB4 expression groups (Figure 5B). TIMER was used to evaluate the link between PFKFB4 expression and purity-regulated immune cell infiltration (B cells, CD4+ T cells, CD8+ T cells, DCs, neutrophils, and macrophages) (Figure 5C). Furthermore, we found that PFKFB4 plays an important role in the immune infiltration of B cells and dendritic cells in LUAD and that B cells and dendritic cells are factors associated with cumulative survival of LUAD (Figure 5D). PDCD-1 and CTLA-4 are important immunological checkpoints for tumor immune evasion. PFKFB4 may be a predictive marker in NSCLC, and we investigated the connection between PFKFB4 and PDCD-1 and CTLA-4 in the TIMER and TCGA databases during our follow-up analysis. PFKFB4 was found to be strongly and positively linked with PDCD-1 and CTLA-4 in patients with NSCLC (Figure 6A and B).
 |
Figure 6 (A and B) Correlation of PFKFB4 expression with PDCD-1 and CTLA-4 expression in NSCLC. |
PFKFB4 Expression-Rich Gene Set
Bioinformatics screening of PFKFB4 co-expressed genes and their functions yielded 981 genes co-expressed with PFKFB4 (Figure 7A and B), and GO analysis showed that these genes were involved in the following biological processes: cell mitosis, cell cycle regulation, cell cycle checkpoint, DNA damage checkpoint, tumor necrosis factor production, regulation of MAP kinase activity, leukocyte migration, lymphocyte migration, lymphocyte aggregation, neutrophil migration, etc. (Figure 7C–H).
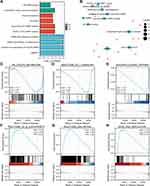 |
Figure 7 (A–B) Enrichment analysis of co-expressed genes. (C–H) GSEA results show differential enrichment between high PFKFB4 expression and low PFKFB4 expression. |
Discussion
PFKFB4 is a member of the PFKFB family, which includes PFKFB1, PFKFB2, PFKFB3, and PFKFB4. PFKFB4 is located on chromosome 3 (region p21 to p22) and encodes an isozyme originally found in testicular tissue that is a key regulator of the glycolytic process.9 The important role in tumorigenesis of the tumor metabolizing enzyme PFKFB4, which regulates glucose metabolism, is reprogrammed in cancer cells. The metabolic underpinning for cancer cell growth and metastasis is increased tumor glycolysis (Warburg effect).10 Patients with high PFKFB4 expression in their breast cancer have a bad prognosis, and it has been established to be an independent breast cancer prognostic factor.11 PFKFB4 is a possible molecular target for gastric cancer because it inhibits the production of oncogenes and assures the proliferation, migration, and invasion of gastric cancer cells.12 PFKFB4 is overexpressed in small cell carcinoma tissues of the prostate, promotes glucose degradation, and maybe an important molecular target.13 one mechanism for the pro-tumor effects of PFKFB4 is its ability to regulate glycolytic flux under normoxic and hypoxic conditions. Furthermore, PFKFB4 has been found to promote tumor progression through a glycolysis-independent mechanism, and enzymes of the core metabolic pathway are important for the development of cancer cells.11 We have demonstrated through immunohistochemical experiments that PFKFB4 is an independent prognostic factor in non-small cell lung cancer. the higher the level of PFKFB4 expression, the worse the prognosis and the more rapid the progression of cancer. The ability of PFKFB4 expression to distinguish tumor from normal tissue and to predict survival at 1, 3, and 5 years suggests that PFKFB4 could be a useful diagnostic and prognostic biomarker for HCC. DNA methylation is a common epigenetic process found in all cancer types.14 Aberrant DNA methylation, which is linked to abnormal gene expression, is one of the hallmarks of cancer. Hypomethylation and activation of metastatic genes are characteristic of cancer cells, and hypomethylation levels of oncogenes can activate high expression of oncogenes, promoting tumor development and suggesting poor patient prognosis.15 We looked into the link between PFKFB4 DNA methylation levels and prognosis in HCC patients. Hypermethylation at 33 CpG sites was linked to a lower chance of survival, with the greatest DNA methylation levels found at cg21141812 and cg207321140.
The immune microenvironment is quite important for tumor development, immunotherapy is a hot topic in today’s society and immunotherapy is helpful in NSCLC.16 Our study suggests that PFKFB4 positively correlates with a variety of immune cells such as B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells in NSCLC. In addition, the biomarkers of these invading immune cells were likewise strongly linked with PFKFB4.17 PFKFB4 may represent the status of the immunological milieu in NSCLC and play a significant role in immune modulation, according to these findings. Immune checkpoint inhibitors are popular cancer treatment, thus we looked into the link between PFKFB4 and immune checkpoints.18 The findings revealed that PFKFB4 expression in NSCLC was positively linked with PD-1 and CTLA-4, implying that inhibiting PFKFB4 could increase the efficacy of immune checkpoint inhibitors in NSCLC.19 PFKFB4 and its associated genes are involved in tumor development, including cell cycle, glycolysis, autophagy, transcriptional regulation, and endothelial tyrosine kinase, according to enrichment analysis.20 Furthermore, via modulating autophagy, PFKFB4 can interact with endothelial tyrosine kinases to control chemoresistance in lung cancer.
However, the biological functions of PFKFB4 in lung cancer cells are still unclear. The proliferation and aggressiveness of tumor cells often determine the prognosis of patients. PFKFB4 is a potential novel molecular target for the treatment of NSCLC, as its overexpression and hypomethylation levels are indicative of poor overall survival and malignant tumor progression, and are mainly involved in the regulation of signaling pathways such as glycolysis and P53.
Conclusion
This suggests that high expression of PFKFB4 in non-small cell lung cancer tissues may contribute to the unlimited proliferation of cancer cells, and therefore inhibition of PFKFB4 expression may be a potential target for the treatment of NSCLC.
Abbreviations
LUAD, lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; TCGA, The Cancer Genome Atlas; OS, Overall Survival; CIBERSORT, Cell type Identification by Estimating Relative Subsets of RNA Transcripts; TMA, Tissue Microarray; AOD, Average Optical Density; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Research Involving Human Participants and/or Animals
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research.
Informed Consent
All patients or their family members provided written informed consent.
Data Sharing Statement
The datasets used and analysed during the current study are available from the corresponding author on reasonable request. NSCLC tissues and matched paraneoplastic tissues, and another TMA (ZL-LUG1201) containing 140 NSCLC tissues was constructed.
Ethical Conduct of Research
The authors state that they have obtained Changzhou Second People’s Hospital Ethics Committee approval and have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Acknowledgments
We acknowledge the TCGA, GEO, TIMER and CIBERSORT databases for free use.
Funding
The following funds support this work:1. “333 Project” of Jiangsu Province (Grant number: BRA2020157)2. “Six One Project”, Research Projects of High-level Medical Personnel of Jiangsu Province (Grant number: LGY2019025)3. High-level Talent Selection and Training Project of the 16th Batch of “Six Talent Peak” in Jiangsu Province (Grant number: WSN-245)4. Medical Scientific Research Foundation of Jiangsu Commission of Health (Grant number: H2018083)5. Jiangsu Provincial Medical Youth Talent (Jiangsu Health Scientific Education 2017 no.3)6. 333 High-Level Talent Training Project (Grant number: 2016, III-0719)7. High-Level Medical Talents Training Project (Grant number: 2016CZBJ042).
Disclosure
All authors declare that they have no competing interests in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi:10.3322/caac.21387
2. Cai YC, Yang H, Shan HB, et al. PFKFB4 overexpression facilitates proliferation by promoting the G1/S Transition and is associated with a poor prognosis in triple-negative breast cancer. Dis Markers. 2021;2021:8824589. doi:10.1155/2021/8824589
3. Chédeville AL, Lourdusamy A, Monteiro AR, et al. Investigating glioblastoma response to hypoxia. Biomedicines. 2020;8:310. doi:10.3390/biomedicines8090310
4. Feng C, Li Y, Li K, et al. Correction to: PFKFB4 is overexpressed in clear-cell renal cell carcinoma promoting pentose phosphate pathway that mediates Sunitinib resistance. J Exp Clin Cancer Res. 2021;40:379. doi:10.1186/s13046-021-02165-5
5. Dasgupta S, Rajapakshe K, Zhu B, et al. Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer. Nature. 2018;556:249–254. doi:10.1038/s41586-018-0018-1
6. Gómez M, Manzano A, Figueras A, et al. Sertoli-secreted FGF-2 induces PFKFB4 isozyme expression in mouse spermatogenic cells by activation of the MEK/ERK/CREB pathway. Am J Physiol Endocrinol Metab. 2012;303:E695–707. doi:10.1152/ajpendo.00381.2011
7. Feng C, Li Y, Li K, et al. PFKFB4 is overexpressed in clear-cell renal cell carcinoma promoting pentose phosphate pathway that mediates Sunitinib resistance. J Exp Clin Cancer Res. 2021;40:308. doi:10.1186/s13046-021-02103-5
8. Pan J-H, Zhou H, Cooper L, et al. LAYN is a prognostic biomarker and correlated with immune infiltrates in gastric and colon cancers. Front Immunol. 2019;10:6. doi:10.3389/fimmu.2019.00006
9. Figueiredo AL, Maczkowiak F, Borday C, et al. PFKFB4 control of AKT signaling is essential for premigratory and migratory neural crest formation. Development. 2017;144:4183–4194. doi:10.1242/dev.157644
10. Gao R, Liu Y, Li D, et al. PFKFB4 promotes breast cancer metastasis via induction of hyaluronan production in a p38-dependent manner. Cell Physiol Biochem. 2018;50:2108–2123. doi:10.1159/000495055
11. Kotowski K, Rosik J, Machaj F, et al. Role of PFKFB3 and PFKFB4 in cancer: genetic basis, impact on disease development/ progression, and potential as therapeutic targets. Cancers. 2021;13:909. doi:10.3390/cancers13040909
12. Gu X, Guan J, Xu J, et al. Model based on five tumour immune microenvironment-related genes for predicting hepatocellular carcinoma immunotherapy outcomes. J Transl Med. 2021;19:26. doi:10.1186/s12967-020-02691-4
13. Kessler R, Fleischer M, Springsguth C, et al. Prognostic value of PFKFB3 to PFKFB4 mRNA ratio in patients with primary glioblastoma (IDH-Wildtype). J Neuropathol Exp Neurol. 2019;78:865–870. doi:10.1093/jnen/nlz067
14. Lu H, Chen S, You Z, et al. PFKFB4 negatively regulated the expression of histone acetyltransferase GCN5 to mediate the tumorigenesis of thyroid cancer. Dev Growth Differ. 2020;62:129–138. doi:10.1111/dgd.12645
15. Meng J, Chen X, Han Z. PFKFB4 promotes lung adenocarcinoma progression via phosphorylating and activating transcriptional coactivator SRC-2. BMC Pulm Med. 2021;21:140. doi:10.1186/s12890-021-01420-x
16. Donnem T, Hald S, Paulsen E, et al. Stromal CD8+ T-cell Density—A promising supplement to TNM staging in non-small cell lung cancer. Clin Cancer Res. 2015;21:2635–2643. doi:10.1158/1078-0432.Ccr-14-1905
17. Wang F, Wu X, Li Y, et al. PFKFB4 as a promising biomarker to predict a poor prognosis in patients with gastric cancer. Oncol Lett. 2021;21:296. doi:10.3892/ol.2021.12557
18. Geng Q, Shen Z, Li L, et al. COL1A1 is a prognostic biomarker and correlated with immune infiltrates in lung cancer. PeerJ. 2021;9:e11145. doi:10.7717/peerj.11145
19. Li Y, Zhao X, Xiao H, et al. APE1 may influence CD4+ naïve T cells on recurrence free survival in early stage NSCLC. BMC Cancer. 2021;21:233. doi:10.1186/s12885-021-07950-1
20. Zhang L, Liu Z, Dong Y, et al. E2F2 drives glioma progression via PI3K/AKT in a PFKFB4-dependent manner. Life Sci. 2021;276:119412. doi:10.1016/j.lfs.2021.119412
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



