Back to Journals » Neuropsychiatric Disease and Treatment » Volume 18
Effect of Early Swimming on the Behavior and Striatal Transcriptome of the Shank3 Knockout Rat Model of Autism
Authors Meng Y , Xu D, Zhang W, Meng W, Lan XY, Wang X, Li M, Zhang X, Zhao Y, Yang H, Zhang R, Zhen Z
Received 18 January 2022
Accepted for publication 17 March 2022
Published 31 March 2022 Volume 2022:18 Pages 681—694
DOI https://doi.org/10.2147/NDT.S357338
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Yuping Ning
Yunchen Meng,1 Dan Xu,1 Weinan Zhang,1 Wenshu Meng,2 Xingyu Lan,3 Xiaoxi Wang,3 Mingjuan Li,3 Xiaoyan Zhang,1 Yu Zhao,1 Haodong Yang,1 Rong Zhang,3– 6 Zhiping Zhen1
1College of P.E and Sports, Beijing Normal University, Beijing, People’s Republic of China; 2College of Life Sciences, Beijing Normal University, Beijing, People’s Republic of China; 3Department of Neurobiology, School of Basic Medical Sciences, Peking University, Beijing, People’s Republic of China; 4Neuroscience Research Institute, Peking University, Beijing, People’s Republic of China; 5Key Laboratory for Neuroscience, Ministry of Education/National Health and Family Planning Commission, Peking University, Beijing, People’s Republic of China; 6Autism Research Center of Peking University Health Science Center, Beijing, People’s Republic of China
Correspondence: Zhiping Zhen; Rong Zhang, Email [email protected]; [email protected]
Background: Autism spectrum disorder (ASD) is a developmental disorder characterized by social behavior deficits and stereotyped behaviors in childhood that lacks satisfactory medical intervention. Early swimming intervention is a noninvasive method combining enriched environment and exercise, which has been proven to improve brain development in young children and to treat neurodevelopmental diseases.
Methods: In this study, we tested the autism-like behavior of rats with deletions in exons 11– 21 of the Shank3 gene and evaluated the effect of early swimming intervention (from postnatal day 8 to 60) on the behavior of this animal model of autism. In addition, the transcriptomes of the striatal tissues of wild-type, Shank3 knockout and Shank3 knockout swimming groups rats were analyzed.
Results: Shank3 knockout rats exhibit core symptoms of autism, and early swimming improved the social and stereotyped behaviors in this autism rat model. Transcriptomics results revealed that compared to the wild-type group, 291 differentially expressed genes (DEGs) were identified in the striatum of the Shank3 knockout group. Compared to Shank3 knockout group, 534 DEGs were identified in the striatum of Shank3 knockout swimming group. The DEGs annotated by Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway shows that the impacts of Shank3 deletion were primarily reflected in synaptic structure, development, morphology, receptor function and signaling, and swimming primarily changed the terms related to the synapses in the striatum of Shank3 knockout rats, including the morphology, structure, composition, development and regulation of synapses.
Conclusion: Early swimming intervention can ameliorate behavioral abnormalities caused by Shank3 knockout, by a mechanism that may involve the process of striatal synaptic development and should be further investigated.
Keywords: Shank3, autism, early swimming, behavior, transcriptome
Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder that occurs before the age of 3 and persists into adulthood.1 Patients with ASD exhibit considerable phenotypic heterogeneity. In addition to the core symptoms of social interaction disorders and repetitive stereotyped behaviors, they also present with symptoms of other mental illnesses, including movement disorders, language impairment, intellectual disability, sensory disorders, developmental delays, sleep disorders, epilepsy, obsessive-compulsive disorder, and attention deficit hyperactivity disorder.2 The prevalence of ASD is high, and patients with ASD require lifelong care.3 ASD imposes a huge economic and mental burden on society and family and has become a major public health problem recognized worldwide.4
The causes of ASD are complex and involve a series of genetic and environmental factors.5 Although the etiology of at least 60% of ASD cases is unclear,6 it is well known that the occurrence of ASD begins during the early stages of brain development and has a lasting impact throughout life. The genetic basis of ASD is complicated. The causative factors involve extensive genetic variation in genetic networks. More than 1000 human ASD risk genes have been reported by the Simons Foundation Autism Research Initiative (SFARI).7 There are many risk genes for ASD, and most of these genes are closely related to the development and plasticity of synapses. These genes or gene products are involved in different aspects of synaptic structures and functions, including regulating the strength or density of synapses, cell adhesion, chromatin remodeling, transcription, cytoskeleton formation and neuron connection.8–10
Shank3 deficiency is one of the most common single-gene risk factors for ASD. In-depth studies of ASD caused by Shank3 deficiency provide a unique opportunity to understand the underlying neuropathological mechanisms of ASD. SHANK3 is a major synaptic scaffold protein that is enriched in excitatory postsynaptic densities and is essential for synapse formation.11 The full length of SHANK3 contains six protein-interacting domains: protein of doubtful function 535 (DUF535); the Ankyrin Repeat domain (ANK); the SRC Homology 3 domain (SH3); the PSD95-DLG1-ZO1 domain (PDZ); the ProlineRich domain (PRO); the Sterile Alpha Motif domain (SAM). Each domain of SHANK3 protein can interact with a variety of synaptic protein molecules and participate in various synaptic functions such as synapse formation, synaptic transmission, and plasticity.12–15 The Shank3 gene was first associated with neurodevelopmental disorders through studies of Phelan-McDermid Syndrome (PMS) and is a key factor inducing the appearance of autistic characteristics in PMS. PMS is a neurodevelopmental disorder caused by the deletion of 22q13.3. In addition to autism-like behaviors (social impairment, stereotyped behaviors), the phenotype is also characterized by hypotonia, language deficits, and brain malformations.15–17 The Shank3 gene is deleted in almost all reported cases of PMS.18–20 Currently, approximately 1% of ASD is attributed to Shank3 gene defects.21,22
Some evidence indicates that multiple symptoms of ASD are closely related to the function of striatal brain regions.23–25 Among them, the dorsal striatum mediates movement and executive function, which are potentially associated with the stereotyped behaviors, repetitive actions, and motor dysfunction observed in ASD patients.26 The nucleus accumbens-striatal pathway primarily mediates reward, cognition, and goal orientation and is related to compulsive behavior, rigid thinking, executive function, and abnormal social coexistence patterns in ASD.27 In the current study on the etiology and intervention mechanism of ASD, we focused more on the striatal brain region.
An enriched environment and physical activity are considered favorable stimuli for neuroplastic processes.28 Swimming combines the multisensory stimulation of an enriched environment with the motor stimulation of physical activity.29 It is one of the most effective exercise programs to promote neurodevelopment and neuroplasticity.30 Proper weight-bearing swimming exercise can have extremely important effects on the nervous system, skeletal muscle and cardiorespiratory function. It plays an important role in maintaining physical and mental health, reducing depression and anxiety, decreasing negative moods, eliminating apprehension, and improving learning and cognition.31 The biggest difference between swimming and other sports is its age appropriateness, which is especially applicable in infants and young children. Many studies have shown that swimming interventions help alleviate the symptoms of autism.32–35
Despite these findings, few studies have focused on the effects and mechanisms of early swimming. Our team has previously demonstrated that Shank3 knockout rats have typical autistic symptoms and have defects in dendritic spine morphology, synaptic protein expression and synaptic transmission in multiple brain regions.36 In this study, we used transcriptomics to explore the effect of Shank3 deletion on striatal gene expression. Further, we studied the effect of early swimming intervention on behavior and on the striatal transcriptome to explore the mechanism of early swimming intervention.
Methods
Generation of Shank3 Rats with Deletion of Exons 11-21
Shank3 gene knockout rats were generously provided by Dr. Zhang Rong, Peking University Health Science Center. Shank3 Δe11-21 rats were generated using CRISPR-Cas9 to delete Shank3 in the germline.36 Subsequent rat genotype identification results were determined by PCR of rat tail DNA using two primers, F1 (CTGTTGGCTGAGCCTGGCATAGAG) and R1 (GCTGGAAAGAAACAACGAGAGCCAG) for the wild-type allele (559 base pairs) and F2 (TTGTGCACTGCCTATGTTGACCACT) and R2 (TAGGCGAGAGAAGATGGTGTGATTTCC) for the mutant allele (688 base pairs).
Animal Husbandry and Care
Sprague-Dawley (SD) rats were housed 2–4 per cage and maintained on a 12:12 h light/dark cycle (indoor temperature 20–25°C, humidity 50–60%) with free access to food and water. All animal experimental procedures conformed to the ethical requirements of Peking University (ethics number ID, LA2015204) and to the National Institutes of Health guidelines for laboratory animal care and use.
Experimental rats were the progeny of Shank3 knockout heterozygous rats breeding pairs (20 pairs), male, 8 days old. According to genotype, young rats were divided into the Shank3 gene knockout group (KO) and littermate wild-type control group (WT). Neonatal rats in the KO and WT groups were randomly selected for swimming training and divided into the Shank3 knockout control group (KC, n=13), the Shank3 knockout swimming group (KS, n=11), the littermate wild-type control group (WC, n=18).
Swimming Exercise
When the pups reached postnatal day (P) 8, the KS group was subjected to swim training for 8 weeks, with 5 days of swimming and 2 days of rest per week. P8-21, rats in the swimming training group were only taken out of the home cage during training, and were still fed by the mothers during the rest of the time. P8 in rats corresponds to approximately 1-year-old children.37 This research study adopted the 8-day-old SD pups’ early swimming training program established by Muniz et al.38 To confirm whether the rat model of ASD could complete the swimming training program, the early swimming intervention program was validated and refined. We extended the period of the training program from P52 to 60, and according to the record of rats’ swimming status, the duration of each training session was increased from 30 min to 40 min at P27-60, as shown in Figure 1. P8-10 was the first week of training, with swimming intervention in a 485×350×200 mm rectangular cage and the water temperature preheated in advance and controlled at 32±1°C. At P8, the rats swam for 2 min, and water submerged the legs of the young rat at approximately 3 cm. At P9, rats swam for 5 min with their stomach submerged in approximately 3.5–4 cm of water. At P10, rats swam for 10 min with the head of the young rat submerged in approximately 4.5–5 cm of water. After P13, a swimming intervention was performed in a circular water tank with a diameter of 150 cm with the water level maintained at 50 cm. During P13-17, daily intervention was performed for 15, 20, 25, 30, 30 min. During P20-24, the rats swam for 30 min a day, and from P27-60, they swam for 40 min a day.
Behavior Testing
At P60, we conducted behavioral tests on each group of rats.
Three-Chamber Test
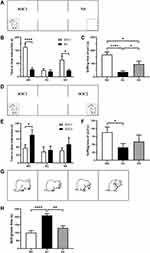
Social behavior was tested in a rectangular box divided into three chambers (40×34×24 cm) with side chambers connected to the middle chamber by a corridor (10×10×15 cm). Rats were tested in the device in the dark, and their activities were recorded by infrared cameras. The test was divided into three phases:39 the adaptation stage, the first stage and the second stage. During the adaptation stage, the rats were allowed to freely explore in the chambers for 5 min. In the first stage (Figure 2A), the social preference test phase, a cage containing an unfamiliar rat of the same sex and age was placed on one side of the device as a social stimulus, and an identical empty cage was placed on the other side. The test rat was placed in the middle chamber and allowed to freely explore the entire device for 10 min. During the second stage (Figure 2D), another unfamiliar rat of the same age and sex was placed in an empty cage as a new social stimulus. The test rat was placed in the middle chamber and allowed to freely explore for 10 min. After each test, the entire apparatus was cleaned using 75% alcohol. The amount of time spent sniffing the wire cage was analyzed by a highly trained observer blinded to the genotype as closely social interaction time.
Self-Grooming Test
Experimental rats were placed inside a rectangular box (40×34×24 cm) for the self-grooming test.36 Infrared cameras were used to record the self-grooming behaviors of rats in the dark. The rats were placed in the box and allowed to adapt for 10 min and then recorded for 10 min. The self-grooming behavior paradigms included rats licking their body and hair, wiping their face with their front paws, and scratching their torso (Figure 2G).
Transcriptomics
Sample Collection and Preparation
After behavioral examination (P63), three striatal tissues of rats in each group were collected for transcriptomics analysis.
RNA quantification and qualification. Trizol was used for RNA extraction. RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
Library preparation for Transcriptome sequencing. Total RNA was used as input material for the RNA sample preparations. mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase, then use RNaseH to degrade the RNA. Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and dNTP. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. To select cDNA fragments of preferentially 370~420 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system.
Data Analysis
Quality Control
Raw data (raw reads) of fastq format were firstly processed through in-house perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing N base and low-quality reads from raw data. At the same time, Q20, Q30 and GC content the clean data were calculated. All the downstream analyses were based on the clean data with high quality.
Reads Mapping to the Reference Genome
Reference genome and gene model annotation files were downloaded from genome website directly. Index of the reference genome was built using Hisat2 v2.0.5 and paired-end clean reads were aligned to the reference genome using Hisat2 v2.0.5. We selected Hisat2 as the mapping tool for that Hisat2 can generate a database of splice junctions based on the gene model annotation file and thus a better mapping result than other non-splice mapping tools.
Differential Expression Analysis
Differential expression analysis of the two conditions/groups (two biological replicates per condition) was performed using the DESeq2 R package (1.20.0). DESeq2 provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P-values were adjusted using Benjamin and Hochberg’s approach for controlling the false discovery rate. Genes with adjusted P-value (PADJ) <0.05 found by DESeq2 and absolute fold change >1 were considered differentially expressed.40
GO and KEGG Enrichment Analysis of Expressed Genes
Gene Ontology (GO) enrichment analysis of differentially expressed genes was implemented using the cluster Profiler R package, in which gene length bias was corrected. GO terms with P-values less than 0.05 were considered significantly enriched differentially expressed genes.
Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database resource for understanding the high-level functions and utility of biological systems, such as cells, organisms and ecosystems from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies (http://www.genome.jp/kegg/). We used the cluster Profiler R package to determine the statistical enrichment of differentially expressed genes in KEGG pathways.41,42
Statistical Analysis
IBM SPSS Statistics 20 (SPSS, Chicago, Illinois, USA) and GraphPad Prism 9.0 (GraphPad Software, San Diego, California, USA) were used for statistical analysis and graph generation. For comparisons, parametric tests, including t-tests and one-way analysis of variance (ANOVA), were used if the data were normally distributed (distribution tested using the Shapiro–Wilk normality test), and nonparametric approaches, including the Wilcoxon test and Kruskal–Wallis test, were used for data with a nonnormal distribution. For all data, the results are expressed as the mean ± standard error of the mean (SEM), and P < 0.05 (two-tailed) was considered statistically significant.
Results
Generation of Shank3-Deficient Rats
Shank3 gene knockout rats were targeted to delete the sequence of exons 11–21 of the Shank3 gene using CRISPR/Cas9 (Figure 3A). The region spanning these two targeted sites was deleted, including the SH3 (SRC homology 3) domain, the PDZ (PSD95-DLG1-ZO1) domain, and the PRO (proline-rich) region. PCR was used to confirm the genotype of these rats (Figure 3B), and transcriptomics results further confirmed the rats’ genotypes (Figure 3C).
Early Swimming Produced an Effect on the Behavior of Rats
Social Preference
Social interaction behaviors and social memory were examined using three-chamber tests. In Phase 1 of the three-chamber tests, the rats were exposed to both social and nonsocial stimuli. Rats in the WC and KS groups spent more time interacting with social partners compared to the empty cage exposure time (WC, P<0.001; KS, P<0.05; Figure 2B). There was no difference in the interaction time between the social partners and the empty cage in the KC group. When comparing the social time of each group, the WC group exhibited significantly more social time than the KC (P<0.001) and KS groups (P<0.05), and the social time of the KS group was significantly higher than for the KC group (P<0.05, Figure 2C). These results indicate that Shank3 knockout rats exhibited social preference deficits, and early swimming repaired this social preference deficit.
Social Memory
In Phase 2 of the three-chamber tests, rats in the WC group displayed a significant social tendency toward new social partners (P<0.05), while rats in the KC group did not have this tendency (Figure 2E). Rats in the KS group spent more time socializing with their new partners than with their old partners, but this difference was not significant. With respect to the interaction time with new social partners among the groups, rats in the WC group exhibited significantly longer interaction time with SOC2 than those in the KC group (P<0.05), but there was no significant difference between the KS group and other groups (Figure 2F). These results indicate that Shank3 knockout rats have social memory deficits. Swimming intervention did not completely reverse this deficiency, but it ameliorated it.
Repetitive Behaviour
Repetitive stereotyped behavior is one of the core symptoms of ASD and manifests as an increase in self-grooming in rodents. Analysis of the duration of self-grooming in the different groups of rats revealed that the duration of self-grooming time of rats in the KC group was significantly longer than in the WC group (P<0.001), and the self-grooming time of the rats in the KS group was significantly lower than that of the KC group (P<0.01, Figure 2H). These observations suggest that Shank3 gene knockout rats exhibit repetitive stereotyped behaviors, and that early swimming intervention reverses this phenotype.
Transcriptome Analysis
To detect the synaptic changes in striatum caused by Shank3 knockout and early swimming intervention, the transcriptomes of the striatal tissues were analyzed.
Differential Gene Analysis
Differential gene analysis identified 291 genes with altered expression patterns in the KC group compared to the WC group, of which 146 genes were upregulated and 145 genes were downregulated (Figure 4A, B and D). The KS group had 143 gene changes, including 89 upregulated genes and 54 downregulated genes (Figure 4A, B and F). In addition, compared to the KC group, there were 534 gene changes in the KS group, of which 351 genes were upregulated and 183 genes were downregulated (Figure 4A, B and E). As shown in Figure 4A, 127 commonly altered genes were identified in the KC vs WC and KS vs KC sets of differential genes, and 42 commonly altered genes were identified in the KC vs WC and KS vs WC sets. The clustering heat map of the differential genes is shown in Figure 4C.
GO Analysis
GO analysis included biological process (BP), cell component (CC), and molecular function (MF). In the GO analysis of differentially expressed genes between the KC and WC groups, we listed the top 20 terms under each category. The major enrichment pathways were related to synapse development, morphology and signal transmission. Among the BP terms (Figure 5A), the autism-related GO terms were central nervous system neuron differentiation (GO:0021953), axonogenesis (GO:0007409), axon development (GO:0061564), regulation of developmental growth (GO:0048638), developmental growth involved in morphogenesis (GO:0060560), myelination (GO:0042552), axon ensheathment (GO:0008366), and central nervous system projection neuron axonogenesis (GO:0021952). In CC terms (Figure 5B), the GO terms associated with autism were main axon (GO:0044304), integral component of presynaptic membrane (GO:0099056), integral component of synaptic membrane (GO:0099699), intrinsic component of presynaptic membrane (GO:0098889), intrinsic component of synaptic membrane (GO:0099240), synaptic membrane (GO:0097060), presynapse (GO:0098793), paranode region of axon (GO:0033270), and dendritic shaft (GO:0043198). MF terms (Figure 5C) primarily reflected changes in a number of ion channels that were closely related to the pathogenesis of autism. These findings suggest that Shank3 deficiency alters synaptic morphology, structure and components in the striatum, leading to autism symptoms.
Between the KS and KC groups, most of the pathways enriched by GO analysis were also related to synaptic function. BP terms (Figure 5D) included axon ensheathment (GO:0008366), regulation of postsynaptic membrane potential (GO:0060078), positive regulation of tissue remodeling (GO:0034105), regulation of tissue remodeling (GO:0034103), positive regulation of neurogenesis (GO:0050769), G-protein coupled receptor signaling pathway (GO:0007188), chemical synaptic and postsynaptic transmission (GO:0099565), and axon ensheathment in central nervous system (GO:0032291). In CC terms (Figure 5E), the structure and function of presynaptic, postsynaptic, synaptic membrane and postsynaptic compact were more reflected. The MF terms (Figure 5F) primarily involved changes in voltage-gated ion channels. These results indicate that early swimming improves synaptic development.
Between the KS and WC groups, there were 4 BP terms (Figure 5G) that were the same as KC vs WC, which were axon development (GO:0061564), axonogenesis (GO:0007409), forebrain development (GO:0030900), telencephalon development (GO:0021537). In CC terms (Figure 5H), there were 5 same pathways as KC vs WC, perikaryon (GO:0043204), mast cell granulen (GO:0042629), extracellular matrix (dendritic shaft), presynapse (GO:0098793). In MF terms (Figure 5I), the same terms as KC vs WC were receptor ligand activity (GO:0048018), receptor regulator activity (GO:0030545), heparin binding (GO:0008201), neuropeptide hormone activity (GO:0005184), hormone activity (GO:0005179). In these common enrichment pathways, the number of differential genes in each term of KS vs WC was less than that of KC vs WC, except for presynapse (GO:0098793). In addition, the unique enrichment pathways in the KS vs WC group were cognition (GO:0050890), memory (GO:0007613), regulation of axonogenesis (GO:0050770), axon guidance (GO:0007411).
KEGG Analysis
KEGG pathway enrichment analysis showed that the differentially enriched pathways between the KC group and the WC group included synaptic receptors and several signaling pathways (Figure 6A). The synaptic receptors were primarily glutamatergic synapses, dopaminergic synapses, cholinergic synapses and GABAergic synapses. The signaling pathways involved included the oxytocin signaling pathway, PI3K-Akt signaling pathway, cGMP-PKG signaling pathway, and MAPK signaling pathway. These enrichment results were closely related to autism. Similarly, pathways that were enriched for differences between the KS and KC groups also included synaptic receptors and several signaling pathways. The synaptic receptors were glutamatergic, cholinergic, GABAergic and dopaminergic. The signaling pathways were the oxytocin signaling pathway and MAPK signaling pathway. The unique terms in KS vs KC were the Ras signaling pathway, calcium signaling pathway, apelin signaling pathway, Rap1 signaling pathway, cAMP signaling pathway, and neurotrophin signaling pathway (Figure 6B). These unique pathways are the result of early swimming intervention and are closely related to neurodevelopment and neuroplasticity. There were 6 same pathways in the KEGG enrichment analysis results between KS vs WC and KC vs WC, which were oxytocin signaling pathway, circadian entrainment, cholinergic synapse, cAMP signaling pathway, axon guidance, MAPK signaling pathway (Figure 6C). The number of differential genes in these terms of KS vs WC was less than that of KC vs WC. In addition, the unique KEGG enrichment pathways in the KS vs WC group were mainly related to glutamatergic and dopaminergic synapses.
Discussion
Similar to our previous research,36 our results demonstrated that Shank3 knockout rats exhibit pronounced social cognitive deficits and repetitive stereotyped behaviors at P60, consistent with the behavioral deficits observed in autism. Early swimming significantly improved these behavioral deficits in Shank3 knockout rats. The striatal transcriptome results revealed possible reasons why Shank3 gene defects cause behavioral disorders and explained the possible mechanism by which early swimming improves behavioral deficits in Shank3 knockout rats. Early swimming may improve the behavior of Shank3 knockout rats by altering presynaptic and postsynaptic structures, receptor function, and neurodevelopment-related signaling pathways.
Epidemiological studies have demonstrated that Shank3 is an important pathogenic gene in autism.43 A large number of studies on animal models with deletions in different exons of the Shank3 gene have demonstrated that Shank3 deletion can cause symptoms of autism.44–49 However, fewer studies have been conducted on Shank3 knockout rat models. Our previous studies demonstrated that rat models with deletion of exons 11–21 of Shank3 exhibit autism-like symptoms.36 In this study, behavioral tests were performed on P60 Shank3 knockout rats, which exhibited more severe autism-like behavior deficits than at P40.
Physical activity and environmental enrichment are considered to be favorable stimuli for neuroplasticity. Swimming combines multiple sensory stimuli in an enriched environment and motor stimulation with physical activity. This is one of the most effective exercise programs for promoting neurodevelopment and neuroplasticity. In previous studies of animal swimming interventions, the intervention time was typically after P21.50–52 However, in our study, we chose P8 through P60 as the swimming intervention time because P8 in rats equates to approximately 1–2 years old in humans, and P60 in rats equates to approximately adulthood in humans. First, we chose this period to correspond to the earliest diagnosis of clinical autism. The earliest appearance of ASD symptoms is 12–24 months after birth.53 More importantly, according to the recommendations of the Centers for Disease Control and Prevention (CDC), early intervention can significantly improve the symptoms associated with autism in children. Treatment of children with ASD should start as soon as the symptoms of ASD appear.
Current studies of swimming interventions for autism focus on people with a target age range of 3–18 years and an intervention period of 8–16 weeks and have assessed the effects of interventions primarily based on motor and social skills. Studies have shown that swimming improves the physical fitness and endurance of children with autism34,54,55 and positively affects cognitive functions, such as social interaction, emotion, and learning.32,35,56 In addition, swimming reduces the occurrence of stereotyped behaviors in children with autism.33,57 We conducted an early swimming intervention study in an animal model of autism and obtained similar results to those of previous authors in terms of behavior. Early swimming improved social interaction, social memory, and repetitive stereotyped behaviors.
The symptoms of autism, especially stereotyped behaviors and movement disorders are closely related to the function of striatal brain regions.44,58,59 Shank3 is most abundantly expressed in the striatum brain region.11 Currently, deletion of the Shank3 gene has been demonstrated to cause structural and partial protein alterations in the striatum.44,46,60 Previous studies on adult Shank3B (exons 13–16 of Shank3 gene were targeted) mice found that the dose of Shank3 had little effect on the changes in directional gene expression in the mouse striatum.61 However, our transcriptome studies in Shank3 knockout rats found different results. GO analysis of the different genes in the striatum of the KC and WC groups revealed that the impacts of Shank3 deletion were primarily reflected in synaptic structure, development, morphology, receptor function and signaling. This may be because we deleted additional exons in Shank3.
Swimming is a rehabilitation training method recommended by the American College of Sports Medicine (ACSM). It has been widely used in the treatment and rehabilitation of diseases, especially in neurological disorders. Previous studies have applied swimming intervention methods to treat animal models of neurological diseases. These studies demonstrated the possible mechanisms by which swimming improves neurological diseases from various aspects, including increased expression of neurotrophic factors in the central nervous system,62–64 changes in synaptic structural proteins,65 changes in synaptic receptor expression,63,66 changes in neurotransmitter release,67 and changes in multiple pathways that affect synaptic plasticity.31,65,68
We observed the same phenomenon in the transcriptomics results of the striatum in the swimming intervention group. We performed GO and KEGG enrichment analyses of differential genes between KS and KC groups. Results demonstrated that swimming primarily changed the terms related to the synapses in the striatum of Shank3 knockout rats, including the morphology, structure, composition, development and regulation of synapses. In addition, KEGG analysis also induced changes in multiple signal pathways, including oxytocin signaling pathway, Ras signaling pathway, calcium signaling pathway, apelin signaling pathway, rap1 signaling pathway, cAMP signaling pathway, and neurotrophin signaling pathway. These signaling pathways are primarily associated with energy metabolism, growth and development, and synaptic plasticity. We also performed differential expression analysis of KS and WC groups and found that the striatum transcriptome of Shank3 knockout rats was closer to that of wild-type rats after the early swimming intervention. Similarly, compared with the KC vs WC group, differential genes were found reduced in the KS vs WC group in the common pathway enriched by GO and KEGG. This indicates that early swimming promotes striatal synaptic development and modulates striatal synaptic signaling through multisensory stimulation and motor stimulation, which in turn ameliorated synaptic abnormalities caused by Shank3 knockout and improved autism-like behavior.
Conclusions
This study demonstrates that a rat model of Shank3 deficiency exhibits symptoms of autism. Autism-like behavior is due to synaptic dysfunction caused by Shank3 gene defects. More importantly, early swimming intervention could reduce stereotyped behaviors and improve social deficits by modulating striatal synaptic plasticity signaling pathways. In the future, we will further explore the potential mechanism of early swimming intervention in improving autism-like behaviors in terms of striatal synaptic morphology and synaptic signaling.
Acknowledgments
We thank Dr. Gao Youhe for advice and comments on the manuscript.
Funding
Supported by Key Project of Beijing Social Science Foundation (No.19YTA007) and Beijing Municipal Science & Technology Commission (No.Z181100001518005).
Disclosure
The authors declare no competing interests in this work.
References
1. Xiao Z, Qiu T, Ke X, et al. Autism spectrum disorder as early neurodevelopmental disorder: evidence from the brain imaging abnormalities in 2–3 years old toddlers. J Autism Dev Disord. 2014;44(7):1633–1640. doi:10.1007/s10803-014-2033-x
2. Hewitson L. Scientific challenges in developing biological markers for autism. OA Autism. 2013;1(1). doi:10.13172/2052-7810-1-1-474
3. Knopf A. Autism prevalence increases from 1 in 60 to 1 in 54: CDC. Brown Univ Child Adolesc Behav Lett. 2020;36(6):4. doi:10.1002/cbl.30470
4. Shahat ARS, Greco G. The economic costs of childhood disability: a literature review. Int J Environ Res Public Health. 2021;18(7):3531. doi:10.3390/ijerph18073531
5. Moog U. The outcome of diagnostic studies on the etiology of mental retardation: considerations on the classification of the causes. Am J Med Genet A. 2005;137a(2):228–231. doi:10.1002/ajmg.a.30841
6. Ellison JW, Rosenfeld JA, Shaffer LG. Genetic basis of intellectual disability. Annu Rev Med. 2013;64(1):441–450. doi:10.1146/annurev-med-042711-140053
7. Lo LHY, Lai KO. Dysregulation of protein synthesis and dendritic spine morphogenesis in ASD: studies in human pluripotent stem cells. Mol Autism. 2020;11(1). doi:10.1186/s13229-020-00349-y
8. O’Roak BJ, Vives L, Girirajan S, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485(7397):246–U136. doi:10.1038/nature10989
9. Iossifov I, O’Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216–U136. doi:10.1038/nature13908
10. Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci. 2015;16(9):551–563. doi:10.1038/nrn3992
11. Monteiro P, Feng GP. SHANK proteins: roles at the synapse and in autism spectrum disorder. Nat Rev Neurosci. 2017;18(3):147–157. doi:10.1038/nrn.2016.183
12. Costales JL, Kolevzon A. Phelan-McDermid syndrome and SHANK3: implications for treatment. Neurotherapeutics. 2015;12(3):620–630. doi:10.1007/s13311-015-0352-z
13. Piao LH, Chen Z, Li QY, et al. Molecular dynamics simulations of wild type and mutants of SAPAP in complexed with Shank3. Int J Mol Sci. 2019;20(1):224. doi:10.3390/ijms20010224
14. Sarowar T, Grabrucker AM. Actin-dependent alterations of dendritic spine morphology in Shankopathies. Neural Plast. 2016;2016:1–15. doi:10.1155/2016/8051861
15. Jaramillo TC, Speed HE, Xuan Z, Reimers JM, Liu S, Powell CM. Altered striatal synaptic function and abnormal behaviour in Shank3 Exon4-9 deletion mouse model of autism. Autism Res. 2016;9(3):350–375. doi:10.1002/aur.1529
16. Kouser M, Speed HE, Dewey CM, et al. Loss of predominant Shank3 isoforms results in hippocampus-dependent impairments in behavior and synaptic transmission. J Neurosci. 2013;33(47):18448–18468. doi:10.1523/jneurosci.3017-13.2013
17. Speed HE, Kouser M, Xuan Z, et al. Autism-associated insertion mutation (InsG) of Shank3 Exon 21 causes impaired synaptic transmission and behavioral deficits. J Neurosci. 2015;35(26):9648–9665. doi:10.1523/Jneurosci.3125-14.2015
18. Gogolla N, Takesian AE, Feng G, Fagiolini M, Hensch TK. Sensory integration in mouse insular cortex reflects GABA circuit maturation. Neuron. 2014;83(4):894–905. doi:10.1016/j.neuron.2014.06.033
19. Filice F, Vörckel KJ, Sungur AÖ, Wöhr M, Schwaller B. Reduction in parvalbumin expression not loss of the parvalbumin-expressing GABA interneuron subpopulation in genetic parvalbumin and shank mouse models of autism. Mol Brain. 2016;9(1):10. doi:10.1186/s13041-016-0192-8
20. Wilson HL, Crolla JA, Walker D, et al. Interstitial 22q13 deletions: genes other than SHANK3 have major effects on cognitive and language development. Eur J Hum Genet. 2008;16(11):1301–1310. doi:10.1038/ejhg.2008.107
21. Delahaye A, Toutain A, Aboura A, et al. Chromosome 22q13.3 deletion syndrome with a de novo interstitial 22q13.3 cryptic deletion disrupting SHANK3. Eur J Med Genet. 2009;52(5):328–332. doi:10.1016/j.ejmg.2009.05.004
22. Durand CM, Betancur C, Boeckers TM, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39(1):25–27. doi:10.1038/ng1933
23. Fuccillo MV. Striatal circuits as a common node for autism pathophysiology. Front Neurosci-Switz. 2016;10. doi:10.3389/fnins.2016.00027.
24. Hollander E, Anagnostou E, Chaplin W, et al. Striatal volume on magnetic resonance imaging and repetitive behaviors in autism. Biol Psychiatry. 2005;58(3):226–232. doi:10.1016/j.biopsych.2005.03.040
25. Li W, Pozzo-Miller L. Dysfunction of the corticostriatal pathway in autism spectrum disorders. J Neurosci Res. 2020;98(11):2130–2147. doi:10.1002/jnr.24560
26. Langen M, Bos D, Noordermeer DS, Nederveen H, van Engeland H, Durston S. Changes in the development of striatum are involved in repetitive behavior in autism. Biol Psychiatry. 2014;76(5):405–411. doi:10.1016/j.biopsych.2013.08.013
27. Kohls G, Schulte-Ruther M, Nehrkorn B, et al. Reward system dysfunction in autism spectrum disorders. Soc Cogn Affect Neur. 2013;8(5):565–572. doi:10.1093/scan/nss033
28. Ball NJ, Mercado E, Orduna I. Enriched environments as a potential treatment for developmental disorders: a critical assessment. Front Psychol. 2019;10. doi:10.3389/fpsyg.2019.00466.
29. Musiyenko O, Chopyk R, Kizlo N. Influence of swimming on sensory functioning, quality of life and behavior of children with autism. Health Sport Rehabil. 2020;6(3):60–69. doi:10.34142/HSR.2020.06.03.07
30. Cheng M, Cong J, Wu Y, et al. Chronic swimming exercise ameliorates low-soybean-oil diet-induced spatial memory impairment by enhancing BDNF-mediated synaptic potentiation in developing spontaneously hypertensive rats. Neurochem Res. 2018;43(5):1047–1057. doi:10.1007/s11064-018-2515-x
31. Aguiar JAS, Castro AA, Moreira EL, et al. Short bouts of mild-intensity physical exercise improve spatial learning and memory in aging rats: involvement of hippocampal plasticity via AKT, CREB and BDNF signaling. Mech Ageing Dev. 2011;132(11–12):560–567. doi:10.1016/j.mad.2011.09.005
32. Pan C-Y. Effects of water exercise swimming program on aquatic skills and social behaviors in children with autism spectrum disorders. Autism. 2010;14(1):9–28. doi:10.1177/1362361309339496
33. Yilmaz I, Yanardağ M, Birkan B, Bumin G. Effects of swimming training on physical fitness and water orientation in autism. Pediatr Int. 2004;46(5):624–626. doi:10.1111/j.1442-200x.2004.01938.x
34. Yanardag M, Akmanoglu N, Yilmaz I. The effectiveness of video prompting on teaching aquatic play skills for children with autism. Disabil Rehabil. 2013;35(1):47–56. doi:10.3109/09638288.2012.687030
35. Ennis E. The effects of a physical therapy-directed aquatic program on children with autism spectrum disorders. J Aquat Phys Ther. 2011;19(1):4–10.
36. Song TJ, Lan XY, Wei MP, et al. Altered behaviors and impaired synaptic function in a novel rat model with a complete Shank3 deletion. Front Cell Neurosci. 2019;13:111. doi:10.3389/fncel.2019.00111
37. Andreollo NA, Santos E, Araújo MR, Lopes LR. Rat’s age versus human’s age: what is the relationship? Arq bras cir dig. 2012;25(1):49–51. doi:10.1590/S0102-67202012000100011
38. de Santana Muniz G, da Silva AM, Cavalcante TC, da Silva França AK, Ferraz KM, Do Nascimento E. Early physical activity minimizes the adverse effects of a low-energy diet on growth and development parameters. Nutr Neurosci. 2013;16(3):113–124. doi:10.1179/1476830512y.0000000037
39. Zhang H-F, Li H-X, Dai Y-C, et al. Electro-acupuncture improves the social interaction behavior of rats. Physiol Behav. 2015;151:485–493. doi:10.1016/j.physbeh.2015.08.014
40. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi:10.1186/gb-2010-11-10-r106
41. Thanseem I, Anitha A, Nakamura K, et al. Elevated transcription factor specificity protein 1 in autistic brains alters the expression of autism candidate genes. Biol Psychiatry. 2012;71(5):410–418. doi:10.1016/j.biopsych.2011.09.020
42. Rose S, Bennuri SC, Davis JE, et al. Butyrate enhances mitochondrial function during oxidative stress in cell lines from boys with autism. Transl Psychiatry. 2018;8(1):1–17. doi:10.1038/s41398-017-0089-z
43. Genovese A, Butler MG. Clinical assessment, genetics, and treatment approaches in Autism Spectrum Disorder (ASD). Int J Mol Sci. 2020;21(13):4726. doi:10.3390/ijms21134726
44. Peca J, Feliciano C, Ting JT, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472(7344):437–U534. doi:10.1038/nature09965
45. Bozdagi O, Sakurai T, Papapetrou D, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1(1):15. doi:10.1186/2040-2392-1-15
46. Wang X, Bey AL, Katz BM, et al. Altered mGluR5-Homer scaffolds and corticostriatal connectivity in a Shank3 complete knockout model of autism. Nat Commun. 2016;7(1):11459. doi:10.1038/ncomms11459
47. Lee J, Chung C, Ha S, et al. Shank3-mutant mice lacking exon 9 show altered excitation/inhibition balance, enhanced rearing, and spatial memory deficit. Front Cell Neurosci. 2015:994. doi:10.3389/fncel.2015.00094.
48. Zhu M, Idikuda VK, Wang J, et al. Shank3-deficient thalamocortical neurons show HCN channelopathy and alterations in intrinsic electrical properties. J Physiol. 2018;596(7):1259–1276. doi:10.1113/jp275147
49. Mei Y, Monteiro P, Zhou Y, et al. Adult restoration of Shank3 expression rescues selective autistic-like phenotypes. Nature. 2016;530(7591):481. doi:10.1038/nature16971
50. Stone V, Kudo KY, Marcelino TB, August PM, Matté C. Swimming exercise enhances the hippocampal antioxidant status of female Wistar rats. Redox rep. 2015;20(3):133–138. doi:10.1179/1351000214Y.0000000116
51. Ma X. Swimming Training Inhibited Inflammation and Cell Apoptosis in Autism Rats. 游泳训练抑制孤独症大鼠炎症反应及细胞凋亡. Genomics Appl Biol. 2020;39(7):3377–3384.
52. Kim J-Y, Yi E-S, Lee H, et al. Swimming Exercise ameliorates symptoms of MOG-induced experimental autoimmune encephalomyelitis by inhibiting inflammation and demyelination in rats. Int Neurourol J. 2020;24(Suppl 1):S39–S47. doi:10.5213/inj.2040156.078
53. Sturner R, Howard B, Bergmann P, et al. Accurate autism screening at the 18-month well-child visit requires different strategies than at 24 months. J Autism Dev Disord. 2017;47(10):3296–3310. doi:10.1007/s10803-017-3231-0
54. Fragala-Pinkham M, Haley SM, O’Neil ME. Group aquatic aerobic exercise for children with disabilities. Dev Med Child Neurol. 2008;50(11):822–827. doi:10.1111/j.1469-8749.2008.03086.x
55. Pan CY. The efficacy of an aquatic program on physical fitness and aquatic skills in children with and without autism spectrum disorders. Res Autism Spect Dis. 2011;5(1):657–665. doi:10.1016/j.rasd.2010.08.001
56. Chu C-H, Pan C-Y. The effect of peer- and sibling-assisted aquatic program on interaction behaviors and aquatic skills of children with autism spectrum disorders and their peers/siblings. Res Autism Spect Dis. 2012;6(3):1211–1223. doi:10.1016/j.rasd.2012.02.003
57. Bumin G, Uyanik M, Yilmaz K, Kayihan H, Topçu M. Hydrotherapy for Rett syndrome. Journal of Rehabilitation Medicine. 2003;35(1):44–45. doi:10.1080/16501970306107
58. Wang WT, Li CC, Chen Q, et al. Striatopallidal dysfunction underlies repetitive behavior in Shank3-deficient model of autism. J Clin Invest. 2017;127(5):1978–1990. doi:10.1172/Jci87997
59. Amodeo DA, Rivera E, Cook EH
60. Yoo T, Cho H, Lee J, et al. GABA neuronal deletion of Shank3 Exons 14-16 in mice suppresses striatal excitatory synaptic input and induces social and locomotor abnormalities. Front Cell Neurosci. 2018;12:341. doi:10.3389/fncel.2018.00341
61. Lee Y, Kang H, Jin C, Zhang Y, Kim Y, Han K. Transcriptome analyses suggest minimal effects of Shank3 dosage on directional gene expression changes in the mouse striatum. Anim Cells Syst. 2019;23(4):270–274. doi:10.1080/19768354.2019.1595142
62. Sigwalt AR, Budde H, Helmich I, et al. Molecular aspects involved in swimming exercise training reducing anhedonia in a rat model of depression. Neuroscience. 2011;192:661–674. doi:10.1016/j.neuroscience.2011.05.075
63. Boracı H, Kirazlı Ö, Gülhan R, Yıldız Sercan D, Şehirli ÜS. Neuroprotective effect of regular swimming exercise on calretinin-positive striatal neurons of Parkinsonian rats. Anat Sci Int. 2020;95(4):429–439. doi:10.1007/s12565-020-00538-y
64. Gyorkos AM, McCullough MJ, Spitsbergen JM. Glial cell line-derived neurotrophic factor (GDNF) expression and NMJ plasticity in skeletal muscle following endurance exercise. Neuroscience. 2014;257:111–118. doi:10.1016/j.neuroscience.2013.10.068
65. Liu W, Xue X, Xia J, Liu J, Qi Z. Swimming exercise reverses CUMS-induced changes in depression-like behaviors and hippocampal plasticity-related proteins. J Affect Disord. 2018;227:126–135. doi:10.1016/j.jad.2017.10.019
66. Ko I-G, Kim S-E, Kim T-W, et al. Swimming exercise alleviates the symptoms of attention-deficit hyperactivity disorder in spontaneous hypertensive rats. Mol Med Rep. 2013;8(2):393–400. doi:10.3892/mmr.2013.1531
67. Leite HR, Mourão FAG, Drumond LE, et al. Swim training attenuates oxidative damage and promotes neuroprotection in cerebral cortical slices submitted to oxygen glucose deprivation. J Neurochem. 2012;123(2):317–324. doi:10.1111/j.1471-4159.2012.07898.x
68. Anand S, Devi SA, Ravikiran T. Differential expression of the cerebral cortex proteome in physically trained adult rats. Brain Res Bull. 2014;104:88–91. doi:10.1016/j.brainresbull.2014.04.012
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.