Back to Journals » Infection and Drug Resistance » Volume 16
Drug Resistance Genes and Molecular Epidemiological Characteristics of Carbapenem-Resistant Klebsiella pneumonia
Authors Liao Y, Gong J, Yuan X, Lu H, Jiang L
Received 12 December 2022
Accepted for publication 6 March 2023
Published 15 March 2023 Volume 2023:16 Pages 1511—1519
DOI https://doi.org/10.2147/IDR.S399142
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Yiqun Liao,1 Junjie Gong,1 Xiaoliang Yuan,2 Hongfei Lu,1 Lixia Jiang1
1Department of Laboratory Medicine, First Affiliated Hospital of Gannan Medical University, Ganzhou, People’s Republic of China; 2Department of Respiratory Medicine, First Affiliated Hospital of Gannan Medical University, Ganzho, People’s Republic of China
Correspondence: Yiqun Liao, Department of Laboratory Medicine, First Affiliated Hospital of Gannan Medical University, 23 Qingnian Road, Zhanggong District, Ganzhou, 341000, Jiangxi Province, People’s Republic of China, Tel +86-15979704263, Email [email protected]
Background: The global prevalence of carbapenem-resistant Klebsiella pneumoniae (CRKP) has become a serious challenge for nosocomial infection and attracted worldwide attention. This study explored the drug resistance genes and molecular characteristics for CRKP, providing a reference for nosocomial prevention and control.
Methods: A total of 42 CRKP isolates were collected from the First Affiliated Hospital of Gannan Medical University (Ganzhou, China) from January 2018 to February 2021. The drug resistance of CRKP was tested by the VitekII Compact system. Drug resistance gene expression was detected by poly-merase chain reaction (PCR), and molecular typing was performed by pulsed-field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST).
Results: All the 42 CRKP isolates were multi-drug resistant. Among them, 35 isolates (83.3%) produced blaKPC-2 and 12 isolates (28.6%) produced blaNDM-1. The detection rate of blaIMP-4 and blaOXA-48 was 2.4% (1/42), respectively. Twelve isolates (28.6%) carried both blaKPC-2 and blaNDM-1, one isolate (2.4%) carried both blaKPC-2 and blaIMP-4, and one isolate (2.4%) carried blaKPC-2, blaNDM-1 and blaOXA-48. A variety of other extended-spectrum beta-lactamases (ESBLs) were also detected. All 42 isolates carried blaSHV and blaCTX-M-1, 27 isolates (64.3%) carried blaTEM and 12 isolates (28.6%) carried blaCTX-M-9. The MLST data classified the 42 CRKP isolates into 11 sequence types, mainly ST11, accounting for 61.9% (26/42), of which 92.3% of isolates (24/26) carrying blaKPC-2. The PFGE results demonstrated that the 42 CRKP isolates could be divided into 20 clusters A-T, with cluster A (26.2%, 11/42) and cluster H (21.4%, 9/42) dominating, which were all ST11.
Conclusion: The CRKP isolates were severely multi-drug resistant, and the main resistant gene was blaKPC-2 production, carrying multiple ESBLs genes simultaneously. The MLST and PFGE revealed that the ST11-blaKPC-2 Klebsiella pneumoniae was the main clonotype. Our findings may offer help to antibiotics selection and nosocomial infection prevention and control.
Keywords: carbapenem-resistant Klebsiella pneumoniae, drug resistance genes, homology, molecular epidemiological characteristics
Introduction
Klebsiella pneumoniae (K. pneumoniae) is one of the most common gram-negative opportunistic pathogens, which can cause respiratory tract infection, urinary tract infection, bloodstream infection and abdominal infection. Carbapenems and other β-lactam antibiotics are normally thought to be a good choice for the treatment of multi-drug resistant gram-negative bacterial infections. But due to the large or unreasonable use of β-lactam antibiotics, extended-spectrum beta-lactamase (ESBLs) and carbapenemase emerge which are capable of hydrolysis of carbapenems. Carbapenem-resistant Klebsiella pneumoniae (CRKP) has emerged subsequently and become a threat to public health, presenting rapid spread over global,1 which can cause higher morbidity and mortality.2,3 There were big differences with the resistance and prevalence of CRKP in different regions of China,4 and the reports from Jiangxi China are limited, which brings great challenges to anti-infection treatment and nosocomial infection control. In our study, we analyzed the homology and the evolutionary relationship between isolates through genotyping and sequencing, and mastered the genome epidemiological characteristics of CRKP, so as to reveal the epidemic trend of CRKP and provide a basis for antibiotics selection, effectively reducing the nosocomial transmission of CRKP.
Materials and Methods
Data Collection
CRKP isolates were those that were resistant to at least one of the carbapenems, including imipenem, meropenem and ertapenem. We collected 42 non-repetitive clinical isolates of CRKP from the First Affiliated Hospital of Gannan Medical University (Ganzhou, China) from January 2018 to February 2021, which all were confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS, Bruker Daltonics, Germany). The patients were composed of 33 males and 9 females, aged from 12 to 91 years old, with an average age of (57.6±18.8) years old.
Antimicrobial Susceptibility Testing (AST)
Forty-two CRKP isolates were tested for antimicrobial susceptibility by the VitekII Compact system (bioMérieux, Marcy l’Etoile, Lyon, France) and the K-B disk diffusion method, with the results based on the standards of the Clinical and Laboratory Standards Institute (CLSI) 2021-M100.5 Quality control strains were Escherichia coli ATCC25922 and K. pneumoniae ATCC700603 (purchased from China National Health Inspection Center).
Detection of Resistance Genes
All of template DNAs were extracted by bacterial DNA Kit (Sangon Biotech, Shanghai, China) and used to detect carbapenemase genes (blaKPC, blaIMP, blaNDM, blaVIM, and blaOXA-48) and ESBLs genes (blaSHV, blaTEM, and blaCTX-M) by PCR. The primers6 are shown in Table 1. The PCR conditions were described as 95°C pre-denaturation for 5 mins, 94°C denaturation for 30s, 55°C annealing for 30s, 72°C extension for 60s, a total of 35 cycles; 72°C extension for 8 min at last. The positive products were sequenced by Shanghai Sangon Biotechnology, and the results were analyzed by BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
 |
Table 1 Primers for Resistance Genes |
Molecular Epidemiological Study
PFGE is a method to separate different-sized restricted fragments of bacterial DNA and analyzes the electrophoresis pattern to determine the strain type according to the typing standard.7 PFGE images were entered into BioNumerics (Version 5.1, Appliedmaths, Inc.) software to identify image bands. Based on the phase between the two images, the similarity coefficient is used for clustering with the unweighted pair group average method (UPGMA) to construct a clustering tree. MLST was performed to explore the genetic correlation of all strains. Seven housekeeping genes (gapA, mdh, pgi, phoE, infB, rpoB, and tonB) were amplified using oligonucleotides representing primer sequences provided by Institute Pasteur MLST (https://bigsdb.pasteur.fr/klebsiella/) (Table 2). The positive products were sequenced by Shanghai Sangon Biotechnology, and the results were submitted to MLST database (https://bigsdb.pasteur.fr/klebsiella/) for comparison.
 |
Table 2 Primer Sequence of K. pneumoniae Housekeeping Genes |
Statistical Analysis
The data were analyzed with SPSS for Windows version 22.0 (SPSS Inc., Chicago, IL, USA).
Results
Patients Data and Distribution of CRKP Isolates
The 42 CRKP isolates mainly originated from the sputum (17/42, 40.5%), followed by bronchoalveolar lavage fluid (8/42, 19.1%), urine (7/42, 16.7%), blood (3/42, 7.1%), and secretion (3/42, 7.1%) (Figure 1A). The patients were composed of 33 males (78.6%) and 9 females (21.4%), aged from 12 to 91 years old, with an average of (57.6±18.8) years old. Of them, 38.1% (16/42) were from intensive care unit (ICU) and 9.5% (4/42) each were from neurosurgery department and VIP department (Figure 1B), of which 70.8% of specimens (17/24) were from respiratory tract.
 |
Figure 1 Distribution of 42 CRKP isolates: (A) source of isolates and (B) departments. |
Antimicrobial Susceptibility Testing (AST)
The 42 isolates demonstrated multi-drug resistance to most of the antimicrobial agents tested, with resistance to amoxicillin-clavulanate, cefuroxime, ceftriaxone, and ceftazidime reaching 100%. The resistance to piperacillin-tazobactam, cefoperazone sulbactam, cefepime, ertapenem, imipenem, meropenem, amikacin, levofloxacin, and trimethoprim-sulfamethoxazole was 88.1%, 97.6%, 97.6%, 92.8%, 83.3%, 90.5%, 21.4%, 61.9%, and 81.0%, respectively. No tigecycline-resistant isolates were detected (Table 3).
 |
Table 3 The Drug Resistance Rate of 42 CRKP Isolates |
Major CRKP Resistance Genes
Among the 42 CRKP isolates, 36 (85.7%) produced carbapenemase, of which 83.3% of isolates (35/42) producing blaKPC-2. 33.3% of isolates (14/42) produced blaNDM, with 12 isolates (28.6%, 12/42) carrying blaNDM-1 and 2 isolates (4.7%, 2/42) carrying blaNDM-5. The detection rate of blaIMP-4 and blaOXA-48 was 2.4% (1/42), respectively. Twelve isolates (28.6%) carried both blaKPC-2 and blaNDM-1, one isolate (2.4%) carried both blaKPC-2 and blaIMP-4, and one isolate (2.4%) carried blaKPC-2, blaNDM-1, and blaOXA-48. The blaVIM gene was not detected in any isolate. A variety of other extended-spectrum beta-lactamases (ESBLs) were also detected. All 42 isolates carried blaSHV, which were mainly blaSHV-12 (61.9%, 26/42). Twenty-seven isolates (64.3%) carried blaTEM, all were blaTEM-1. All isolates carried blaCTX-M-1, mainly blaCTX-M-55 (59.5%, 25/42), and 12 isolates (28.6%) carried blaCTX-M-9, mainly blaCTX-M-65 (19.1%, 8/42) (Table 4). Some of the sequencing maps are shown in Figure 2.
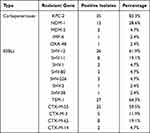 |
Table 4 Resistance Genes of 42 CRKP Isolates |
 |
Figure 2 Sequencing map of blaKPC-2. |
Molecular Typing Based on MLST and PFGE
The MLST results showed that 42 CRKP isolates were divided into 11 sequence types, which were mainly ST11, accounting for 61.9% (26/42), of which 92.3% of isolates (24/26) carrying blaKPC-2. The remaining ST types were ST307 (5 strains), ST43 (2 strains), ST476 (2 strains), ST831 (1 strain), ST27 (1 strain), ST327 (1 strain), ST290 (1 strain), ST15 (1 strain), ST73 (1 strain), ST147 (1 strain). The 42 CRKP isolates were divided into 31 PFGE types, and 20 PFGE clusters A-T at the level of 85% similarity coefficient,7 with cluster A (26.2%, 11/42) and cluster H (21.4%, 9/42) dominating. The clonal subtypes of A1, A3, and H6 were all ST11. However, 26 PFGE types contained only 1 isolate, that is, each of the 26 isolates had a unique PFGE type (Figure 3).
 |
Figure 3 PFGE analysis. |
Discussion
Carbapenems including imipenem and meropenem are usually used as effective antibiotics in the treatment of severe infections caused by multi-drug resistant K. pneumoniae. However, data from CARSS (China Antimicrobial Resistance Surveillance System) showed the resistance rate of K. pneumoniae to imipenem and meropenem has rapidly increased from 3.0% and 2.9% in 2005 to 25.0% and 26.3% in 2018.8 The number of CRKP isolates also has increased rapidly in the past decade. In China and the European countries, CRKP accounted for about 70%~90% of carbapenem-resistant enterobacteria (CRE).9,10 In this study, the CRKP mainly isolated from sputum and bronchoalveolar lavage fluid, and the patients were mainly from ICU and neurosurgery, who had been treated with numerous antibiotics. The study showed that the CRKP isolates were extensively drug resistant, with resistance to third and fourth-generation cephalosporins highly reaching 100%, and the resistance to imipenem and meropenem were 83.3% and 90.5%, respectively. There were relatively consistent with the situation in Zhejiang, Guangdong, Sichuan China,11–13 which extremely limited antibiotics selection in clinic. Clinical laboratories should jointly carry out more antibiotics sensitivity tests, including fosfomycin, chloramphenicol and sulbactam, etc, in order to find possible effective anti-infective antimicrobials, which are helpful to the accurate use of antibiotic and the prevention and control of CRKP infection in hospitals.
The resistance mechanisms mainly include producing carbapenemase, extended-spectrum beta-lactamases (ESBLs) or AmpC enzymes combined with structural mutations, such as the lack of outer membrane proteins and high expression of effluence pump genes or changes in penicillin-binding proteins.14 At present, carbapenemase production is the main mechanism and blaKPC-2 is the most common resistance gene in K. pneumoniae in China.10,15 Since the first report of K. pneumoniae producing blaKPC-2 carbapenemase in Zhejiang China in 2007, CRKP has been reported all over the country, such as Beijing, Shanghai, Jiangsu, etc.16 Outbreaks of CRKP producing blaKPC-2 carbapenemase also have been reported in the United States, Italy and Spain.17,18 Our study depicted that 85.7% of isolates produced carbapenemase, of which blaKPC-2 was detected in 83.3% of isolates, indicating that blaKPC-2 production was the main resistant gene in our hospital, which was consistent with the research report that blaKPC-2 is the main carbapenemase genotype carried by CRKP in China. blaNDM-1 is a metallo-β-lactamase with strong hydrolytic activity in Enterobacter commonly, which can hydrolyze penicillin, cephalosporin, cephamycin, and other β-lactam antimicrobials. Isolates carrying blaNDM-1 are multi-drug resistant, and it has been reported that blaNDM-1 can be spread horizontally between different bacterial species through plasmids and integrons,19 becoming a “super bacteria”. In our study, the detection rate of blaNDM-1 was 28.6%, it also indicated the widespread of blaNDM-1 in our hospital, which was consistent with that blaNDM-1 was the main genotype of CRKP in Hainan province, China,20 but quite different from the reports that the detection rate of blaNDM was only 1.7% in Changsha China6 and Shanghai China.21 So the present study indicated that there were regional differences in the prevalence of carbapenemase genes. Therefore, it is necessary to conduct multi-center and transverse investigations of drug resistance genes over the country.
Carbapenemase production isolates also carried multiple ESBLs (blaSHV, blaTEM, blaCTX-M, etc.) simultaneously. Our study revealed that 61.9% and 64.3% of isolates carried blaSHV-12 and blaTEM-1 respectively, which was consistent with domestic research and abroad.22,23 The subtypes of blaCTX-M were mainly blaCTX-M-55 (59.5%) and blaCTX-M-65 (19.1%), which was quite different with the results from Shandong Province and Beijing China,15,22 it may be related to clinical medication habits, the number of isolates and different regions. There were 64.3% of isolates carrying three types of ESBLs simultaneously, suggesting that the coexistence of multiple resistance genes may be an important cause of the extensive drug resistance of CRKP in our hospital.
The PFGE results showed that the 42 CRKP isolates were divided into 31 PFGE types and 20 PFGE clusters A-T, with cluster A (26.2%) and cluster H (21.4%) dominating; however, 26 isolates had a single and unique PFGE type, indicating the polymorphism and the complexity of the genetic background in our hospital. The result of MLST showed that 26 isolates (61.9%) were the dominant ST11 type, of which 92.3% of isolates (24/26) carrying blaKPC-2, which was consistent with the domestic research result.24 ST11 and ST258 belong to the CC258 clonal complex, with only one site difference between them. ST258 was less reported in China, also not found in our study, but experienced large-scale outbreaks in the United States and other countries,4,25 which indicated there were differences in prevalence in China and abroad.
Conclusion
In summary, the CRKP in our hospital is resistant to most of common antibiotics and presents multi-drug resistance, so antibiotics selection needs to be rationalized and standardization. Mechanism of carbapenem resistance is related to different resistant genes, with blaKPC-2 dominating and blaNDM-1 needing to be focused on. ST11 is the main clonotype of CRKP. So ST11-blaKPC-2 Klebsiella pneumoniae extremely needed to be focused on clinical management and effective measures to prevent and control further dissemination. Therefore, it is necessary to strengthen the antibiotic resistance monitoring and conduct epidemiological investigation termly.
Ethics and Consent Statement
All the isolates were part of the routine hospital laboratory procedure, and no samples were collected specifically for this research. This study has been designed in accordance with the Declaration of Helsinki (2013) and approved by the First Affiliated Hospital of Gannan Medical University.
Funding
This study is supported by Science and Technology Project of Health Commission of Jiangxi, China (No. 202130651) and Science and Technology Project, Department of Education, Jiangxi, China (No. GJJ201534).
Disclosure
All authors have no conflicts of interest in this work.
References
1. Hu Y, Liu C, Shen Z, et al. Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008–2018. Emerg Microbes Infect. 2020;9(1):1771–1779. doi:10.1080/22221751.2020.1799721
2. Li M, Wang X, Wang J, et al. Infection-prevention and control interventions to reduce colonisation and infection of intensive care unit-acquired carbapenem-resistant Klebsiella pneumoniae: a 4-year quasi-experimental before-and-after study. Antimicrob Resist Infect Control. 2019;8(1):8. doi:10.1186/s13756-018-0453-7
3. Vardakas KZ, Matthaiou DK, Falagas ME, et al. Characteristics, risk factors and outcomes of carbapenem-resistant Klebsiella pneumoniae infections in the intensive care unit. J Infect. 2015;70(6):592–599. doi:10.1016/j.jinf.2014.11.003
4. Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(suppl_1):S28–S36. doi:10.1093/infdis/jiw282
5. Clinic and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: M100-S31[S]. Wayne, PA: CLSI; 2021.
6. He H, Huang ZY, Tao XY, et al. Mechanism of carbapenemase-producing and homology analysis of carbapenem-resistant Klebsiella pneumoniae. Chin J Nosocomiol. 2021;31(03):326–331.
7. Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction pattern produced by pulse-field gel electrophoresis criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi:10.1128/jcm.33.9.2233-2239.1995
8. Hu FP, Guo Y, Zhu DM, et al. CHINET surveillance of bacterial resistance: results of 2020. Chin J Infect Chemother. 2021;21(4):377–387.
9. Grundmann H, Glasner C, Albiger B, et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis. 2017;17(2):153–163. doi:10.1016/S1473-3099(16)30257-2
10. Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi:10.1016/j.ebiom.2017.04.032
11. Ding YP, Lu J, Li X, et al. Analysis of drug resistance genes in carbapenem-resistant K. pneumoniae based on WGS. Chin J Clin Infect Dis. 2019;12(02):122–126.
12. Huang JM, Ke BX, He DM, et al. Drug resistance and molecular epidemiological characteristics of carbapenem-resistant Klebsiella pneumoniae in Guang dong. Chin J Nosocomiol. 2022;32(06):813–818.
13. Liu L, Feng Y, Tang G, et al. Carbapenem-resistant isolates of the Klebsiella pneumoniae complex in Western China: the common ST11 and the surprising hospital-specific types. Clin Infect Dis. 2018;67(suppl_2):S263–S265. doi:10.1093/cid/ciy662
14. Zhang P, Shi Q, Hu H, et al. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin Microbiol Infect. 2020;26(1):124.e1–124.e4. doi:10.1016/j.cmi.2019.08.020
15. Chi X, Hu G, Xu H, et al. Genomic analysis of A KPC-2-producing Klebsiella Pneumoniae ST11 outbreak from a teaching hospital in Shandong Province, China. Infect Drug Resist. 2019;12:2961–2969. doi:10.2147/IDR.S221788
16. Wang Q, Wang X, Wang J, et al. Phenotypic and genotypic characterization of carbapenem-resistant enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis. 2018;67(suppl_2):S196–S205. doi:10.1093/cid/ciy660
17. Lee CR, Lee JH, Park KS, et al. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. doi:10.3389/fmicb.2016.00895
18. Oteo J, Pérez-Vázquez M, Bautista V, et al. The spread of KPC-producing Enterobacteriaceae in Spain: WGS analysis of the emerging high-risk clones of Klebsiella pneumoniae ST11/KPC-2, ST101/KPC-2 and ST512/KPC-3. J Antimicrob Chemother. 2016;71(12):3392–3399. doi:10.1093/jac/dkw321
19. Gao H, Liu Y, Wang R, et al. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine. 2020;51:102599. doi:10.1016/j.ebiom.2019.102599
20. Li TJ, Wu H, Wang MX, et al. Drug resistance genes in carbapenem-resistant Klebsiella pneumoniae isolated from ICU and their homology. Chin J Nosocomiol. 2021;31(17):2658–2662.
21. Li XJ, Ma W, Guo J, et al. Epidemiological characteristics and molecular biology of carbapenem-resistant Klebsiella pneumoniae. Acad J Sec Mil Med Univ. 2020;41(10):1109–1114.
22. Wang Q, Li Y, Zheng B. Antimicrobial susceptibility and resistant genes of carbapenem-resistant Klebsiella pneumoniae in China from 2017 to 2018. Chin J Infect Control. 2021;20(05):437–442.
23. Bhaskar BH, Mulki SS, Joshi S, et al. Molecular characterization of extended spectrum β-lactamase and carbapenemase producing Klebsiella pneumoniae from a tertiary care hospital. Indian J Crit Care Med. 2019;23(2):61–66. doi:10.5005/jp-journals-10071-23118
24. Yu F, Lv J, Niu S, et al. Multiplex PCR analysis for rapid detection of Klebsiella pneumoniae carbapenem-resistant (sequence type 258 [ST258] and ST11) and hypervirulent (ST23, ST65, ST86, and ST375) strains. J Clin Microbiol. 2018;56(9):e00731–18. doi:10.1128/JCM.00731-18
25. Castanheira M, Deshpande LM, Mills JC, et al. Klebsiella pneumoniae isolate from a New York City hospital belonging to sequence type 258 and carrying blaKPC-2 and blaVIM-4. Antimicrob Agents Chemother. 2016;60(3):1924–1927. doi:10.1128/AAC.01844-15
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
