Back to Journals » Journal of Inflammation Research » Volume 15
Doxorubicin-Induced Cardiotoxicity May Be Alleviated by Bone Marrow Mesenchymal Stem Cell-Derived Exosomal lncRNA via Inhibiting Inflammation
Authors Tian C, Yang Y, Li B, Liu M, He X, Zhao L, Song X, Yu T, Chu XM
Received 14 January 2022
Accepted for publication 9 July 2022
Published 6 August 2022 Volume 2022:15 Pages 4467—4486
DOI https://doi.org/10.2147/JIR.S358471
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Chao Tian,1,* Yanyan Yang,2,* Bing Li,3 Meixin Liu,4 Xiangqin He,4 Liang Zhao,4 Xiaoxia Song,4 Tao Yu,4,5 Xian-Ming Chu1,6
1Department of Cardiology, The Affiliated Hospital of Qingdao University, Qingdao, People’s Republic of China; 2Department of Immunology, Basic Medicine School, Qingdao University, Qingdao, People’s Republic of China; 3Department of Genetics, Basic Medicine School, Qingdao University, Qingdao, People’s Republic of China; 4Department of Cardiac Ultrasound, The Affiliated Hospital of Qingdao University, Qingdao, People’s Republic of China; 5Institute for Translational Medicine, The Affiliated Hospital of Qingdao University, Qingdao, People’s Republic of China; 6Department of Cardiology, The Affiliated Cardiovascular Hospital of Qingdao University, Qingdao, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Tao Yu, Institute for Translational Medicine, The Affiliated Hospital of Qingdao University, Qingdao, People’s Republic of China, Tel/Fax +86-532-82991791, Email [email protected] Xian-Ming Chu, Department of Cardiology, the Affiliated Hospital of Qingdao University, No. 59 Haier Road, Qingdao, 266100, People’s Republic of China, Tel +86-532-82913257, Email [email protected]
Purpose: To explore the therapeutic mechanism of bone marrow mesenchymal stem cells derived exosomes (BMSC-Exos) for doxorubicin (DOX)-induced cardiotoxicity (DIC) and identify the long noncoding RNAs’ (lncRNAs’) anti-inflammation function derived by BMSC-Exos.
Materials and Methods: High-throughput sequencing and transcriptome bioinformatics analysis of lncRNA were performed between DOX group and BEC (bone marrow mesenchymal stem cells derived exosomes coculture) group. Elevated lncRNA (ElncRNA) in the cardiomyocytes of BEC group compared with DOX group were confirmed. Based on the location and co-expression relationship between ElncRNA and its target genes, we predicted two target genes of ElncRNA, named cis_targets and trans_targets. The target genes were analyzed by enrichment analyses. Then, we identified the key cellular biological pathways regulating DIC. Experiments were performed to verify the therapeutic effects of exosomes and the origin of lncRNAs in vitro and in vivo.
Results: Three hundred and one lncRNAs were differentially expressed between DOX and BEC groups (fold change > 1.5 and p < 0.05), of which 169 lncRNAs were elevated in the BEC group compared with the DOX group. GO enrichment analysis of target genes of ElncRNAs showed that they were predominantly involved in inflammation-associated processes. KEGG analysis indicated that their regulatory pathways were mainly involved in oxidative stress-induced inflammation and proliferation of cardiomyocyte. The verification experiments in vitro showed that the oxidative stress and cell deaths were decreased in BEC groups. Moreover, from the top 10 ElncRNAs identified in the sequencing results, MSTRG.98097.4 and MSTRG.58791.2 were both decreased in the DOX group and elevated in BEC group. While in verification experiments in vivo, only the expression of MSTRG.58791.2 is consistent with the result in vitro.
Conclusion: Our results show that ElncRNA, MSTRG.58791.2, is possibly secreted by the BMSC-Exos and able to alleviate DIC by suppressing inflammatory response and inflammation-related cell death.
Keywords: doxorubicin-induced cardiotoxicity, exosomes, inflammation, transcriptome sequencing analysis
Introduction
The rapid development of immune checkpoint inhibitors and targeted therapy has advanced tumor therapy into a new era of precision medicine. Chemotherapy remains the cornerstone of oncological treatment. Doxorubicin (DOX), a variety of cross-sectional anthracycline, is indispensable in chemotherapy. However, it can also induce senescence,1 apoptosis,2 pyroptosis,3 autophagy,4 and ferroptosis5 of cardiomyocytes, causing arrhythmia, hypertrophic cardiomyopathy, and congestive heart failure while killing tumor cells.6 The cardiovascular damage associated with DOX chemotherapy, termed doxorubicin-induced cardiotoxicity (DIC), seriously threatens the health of cancer patients due to its progressive, irreversible nature.7 At present, dexrazoxane (DZR) is recognized as the only drug to prevent DIC, but it has a limited effect and poses the risk of secondary tumors or bone marrow suppression.8 Therefore, there is a need to find potential biomarkers and elucidate the molecular mechanism of DIC, which will benefit the exploration of effective targets for solving the trouble.
It has been proven that DOX triggers the production of ROS, which result in apoptosis, lipid peroxidation of myocardial cell membranes and damage to myocardial mitochondrial DNA.9,10 Reducing ROS levels in cardiomyocytes may be an effective defense against DIC. However, it remains unclear whether ROS is the only significant factor in DIC. Inflammation and related cell death (apoptosis or pyroptosis) may also play a role.
In recent years, exosomes have garnered much attention. These vesicles are released into body fluid by all cells and can regulate recipient cells’ biological activities by delivering nucleic acids, including DNA, messenger RNAs, microRNAs, long noncoding RNA (lncRNAs), circular RNA, transfer RNA, and piwi-interacting RNAs.11,12 These noncoding RNAs were also used as therapeutic targets for various cardiovascular diseases. Furthermore, exosomes lack immunogenicity due to their outer biological membrane. Given these merits, they have been considered to be a latent therapeutic strategy for cardiovascular diseases,13–19 autoimmune diseases,20 as well as DIC.21 For example, researchers have explored exosomes derived from embryonic stem cells to curb DOX-induced pyroptosis.22,23
Long noncoding RNAs (lncRNAs) have been involved in the apoptosis,24 proliferation or regeneration of cardiomyocyte.25–27 Moreover, lncRNAs in exosomes have been shown to be involved in the inhibition of cardiomyocyte apoptosis28,29 and pyroptosis.30
In this study, we constructed DIC model in vitro firstly. Then, transcriptome sequencing of lncRNA was performed between DOX group and BEC group, which confirmed the therapeutic effect of bone marrow mesenchymal stem cell-derived exosome (BMSC-Exos) on DIC combined with experiments in vitro and in vivo. At last, we discussed and verified whether elevated lncRNA (ElncRNA) was from BMSC-Exo or not.
Materials and Methods
Preparation of Samples for Sequencing
Cell Culture
Using Kunming mice at 1‒3 days postnatally, we isolated primary neonatal cardiomyocytes as described previously.31,32 These primary cardiomyocytes were cultured in DMEM/F-12 medium (Gibco, Amarillo, TX, USA) supplemented with 5% fetal bovine serum (FBS) (ExCell Bio, Shanghai, China). Primary BMSCs (Procell Life Science & Technology Co., Ltd, Wuhan, China) were cultured in conditioned medium supplemented with 10% exosome free-FBS (System Biosciences, Palo Alto, CA, USA) for coculture with DOX-injured cardiomyocytes in order to eliminate the interference of exosomes in FBS.
Cell Groups and Coculture
Two groups of cardiomyocytes were prepared. In the DOX group, primary cardiomyocytes were exposed to 2 μmol/L DOX (Sigma Aldrich, USA) for 24 hours. Then, the culture medium was replaced with fresh medium (medium supplemented with 5% FBS) for 48 hours. The experiment was repeated three times (DOX1, DOX2, DOX3). In the BEC group, DOX-injured cardiomyocytes prepared as for the DOX group were cocultured with BMSCs in a Transwell coculture system (Corning, New York, USA) (BEC1, BEC2, BEC3), as previously described.33 The pore diameter of Transwell is 0.4μm, which is considered to permit the transmit of exosomes derived from the cells in the upper chambers. After 48 hours of coculturing, total RNA of cardiomyocytes both in the DOX group and BEC group was extracted using SparkZol (SparkJade, Jinan, China) for sequencing.
Bioinformatics Analysis
cDNAs reversely transcribed from the total RNAs in the DOX and BEC were sequenced by an Illumina HiSeq high-throughput sequencing platform (San Diego, CA, USA) based on sequencing by synthesis (SBS). Raw read data in the FASTQ format were firstly processed through Perl scripts. Clean reads were obtained by removing reads containing adapter and other components without regulatory functions from the raw data. All the downstream analyses were based on clean data. The genome Mus musculus (GRCm38_release95. Mus_musculus. GRCm38_release95.genome.fa) was employed as the reference database. The transcriptome was assembled using the String Tie software (v1.3.1) based on the reads mapped to the reference genome after obtaining clean sequencing data. We obtained mapped data that predicted alternative splicing, structure, and new genes based on sequence comparisons. We annotated the assembled transcripts by means of the gffcompare program (0.9.8). CPC2 (CPC2-beta),34 CNCI (v2),35 Pfam (v1.3),36 and CPAT (1.2.2)37 software programs were combined to sort non-protein coding RNA candidates. Moreover, they were used to select lncRNA candidates.
Quality control, including the classification, expression, and sequence conservation of lncRNAs, was conducted before the differential expression analysis. Cuffcompare (v2.1.1) software was wielded to confirm the different classifications of lncRNAs. String Tie was used to predict the expression level of lncRNAs, which was indicated by the fragments per kilobase of transcript per million fragments mapped. Conservation between the lncRNA and mRNA was compared by means of the phastCons software.38
LncRNAs that were differentially expressed (DE_lncRNAs) between the DOX and BEC group were identified and visualized as a heat map by R package. Pearson’s Correlation Coefficient was used to assess the intra-class differences (DOX1 vs DOX2 vs DOX3, and BEC1 vs BEC2 vs BEC3) and the inter-class differences (DOX vs BEC). The smaller the differences within a group and the larger the difference between the groups, the more significant was the difference. We set the parameter of log2FC (FC, fold change) >1.5 and p-value <0.05 as the selective standard for defining differential expression of lncRNAs between the DOX and BEC groups. We focused on identifying the elevated lncRNAs (ElncRNAs) in the cardiomyocytes in the BEC group, which were regarded to be secreted by BMSC-Exos in the upper chamber. But it needs evidence combined with bioinformatic analyses to support this hypothesis.
Prediction of the Targets of ElncRNAs and Their Enrichment Analyses
Based on the mode of interaction of lncRNAs and their target genes, we adopted two different prediction methods. The first was mainly based on the positional relationship between the lncRNA and the target genes. Neighboring genes, within a 100-kb range, were considered as cis_target genes. The second was based on the correlation of lncRNA and mRNA expression levels between samples. The target genes predicted by this method were defined as trans_target genes. Specifically, the Pearson correlation coefficient was used as the indicator for the correlation between lncRNA and mRNA among samples, and the genes with correlation absolute value >0.9 and P value <0.01 were selected as the trans_target genes of lncRNA.
The differences in target gene clusters were determined by MCODE software. ClusterProfiler39 was used for Gene Ontology (GO) analysis to reveal the cellular component (CC), biological process (BP), and molecular function (MF) of the genes targeted by ElncRNAs. KOBAS software was employed to test enrichment of differentially expressed genes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. In addition, a novel KEGG enrichment analysis based on the pathway types developed by Biomarker Technologies was adopted. STRING40 (version 11.0) was implemented to predict and validate protein‒protein interactions. The constructed protein interaction network was imported into Cytoscape41 software for visualization.
Cell Verification Experiments
Cardiomyocyte Viability Detection
Three groups of cardiomyocytes were prepared, cardiomyocytes without any treatment (NC group), DOX group and BEC group. Their viability was detected by using a Cell Counting Kit-8 (Meilunbio, Dalian, China) according to the instructions. The OD value of different groups was used as the index of viability.
Reactive Oxygen Species (ROS) Detection
As the increase of oxidative stress is a feature for DIC, we confirmed whether BMSC-Exo can decrease it to alleviate DIC by ROS detection. A Reactive Oxygen Species Assay Kit (Solarbio, Beijing, China), based on a 2’-7ʹdichlorofluorescin diacetate fluorescent probe, was used for detection of ROS. First, we loaded the probe in three groups of cardiomyocytes, with the fluorescent probe by incubation according to the manufacturer’s instructions. After washing the cells, the samples were viewed directly under a laser confocal microscope. An excitation wavelength of 488 nm and emission wavelength of 525 nm were used to detect the fluorescence intensity in real time. Image J was utilized for quantitative analysis based on fluorescence intensity.
Real-Time Quantitative Polymerase Chain Reaction (qRT-PCR) for Cell and Animal Verification Experiments
To confirm the results obtained by sequencing, the top-10 ElncRNAs, we performed real-time quantitative polymerase chain reaction (qRT-PCR). Total RNA was extracted from cell and heart tissue samples using SparkZol reagent. For mRNA detection, cDNA was reverse transcribed by using a Hifair III 1st Strand cDNA Synthesis kit (YEASEN Biotechnology, Shanghai, China). A Hieff qPCR SYBR Green Master Mix (YEASEN Biotechnology) was used for quantification. GAPDH was employed as the control to evaluate the expression level of lncRNAs and inflammation markers. 2−ΔΔCt method was utilized to calculate the quality of mRNA. The primer sequences are presented in Table 1.
 |
Table 1 The Primer Sequence for qRT-PCR |
Animal Experiments
The Isolation of Exosomes
We have utilized the differential ultracentrifugation method to isolate exosomes from BSMC.42 In brief, the culture medium of BMSC was collected and subjected to differential centrifugation steps. Firstly, they were centrifuged at 300×g for 10 min, we collected the supernatant. Next, the supernatant was subjected to 2000×g for 15 min to remove floating and dead cells. The supernatants were further centrifuging at 10,000×g for 30 min to remove large extracellular vesicles. At last, exosomes were isolated from the supernatant by ultracentrifugation at 100,000×g for 2h at 4°C. We used 200μL ice-cold PBS to resuspend the exosomes. The concentration of exosomal proteins was quantified by BCA Protein Assay Kit (YEASEN Biotechnology, Shanghai, China).
Identification of Exosomes by Electron Microscopy
20μL suspension of exosome was absorbed by pipetting gun, then dropped into the copper mesh grid under electron microscope, stood for about 2 minutes, and fixed exosomes with 2% phosphotungstate solution for about 5 minutes. After the excess water was absorbed by filter paper, the exosomes were dried naturally at room temperature. Exosomes were observed under the transmission electron microscope (TEM) and photographed.
Identification of Exosomes by NTA
The sample pool was cleaned with deionized water, and the NTA instrument was calibrated with polystyrene microspheres (100 nm). The sample pool was cleaned with PBS. And the sample was diluted 1000 times to 108/mL. The concentration of exosomes in the supernatant of BMSC was determined first, and then the diameter size distribution was determined.
The Establishment of Animal Model
Thirty male C57BL/6 mice aged 10 weeks (20–25g) were randomly divided into 3 groups: normal control (NC) group, DOX treatment (DOX) group and DOX+BMSC-Exosome treatment (DOX+BMSC-Exo) group. In NC group, mice were fed normally without any treatment; in DOX group, mice were intraperitoneally injected with doxorubicin on day 1 (10 mg/kg) and day 8 (10 mg/kg), and acute doxorubicin-induced cardiotoxicity (DIC) model was established in vivo at a cumulative dose of 20 mg/kg. In DOX+BMSC-Exo group, 50μg BMSC-Exosomes quantized by BCA assay were injected into the mice via tail vein on day 8, day 11 and day 14, respectively.
The Sirius Red Staining of Heart Tissues
The samples of Murine heart tissue were fixed with 4% paraformaldehyde firstly, and then they were embedded into paraffin. Tissue sections were then subjected to 3 mm thick for Sirius Red staining.
Statistical Analysis
Image J (v1.8.0) software (NIH, Bethesda, MD, USA) was used to analyze the level of fluorescence in the ROS detection experiment and to set the scale bar. GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA, USA) was utilized to conduct statistical analysis and generate graphs. The unpaired Student’s t-test was applied to assess statistical significance of differences between DOX and NC group or BEC and DOX group. A p-value <0.05 was regarded significant. For Kaplan-Meier Curve, the p-value was used for the Log-rank (Mantel-Cox) test.
Results
Quality Control of lncRNAs
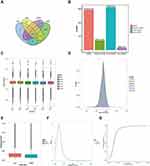
In total, 34,874 lncRNAs were predicted from the cDNAs of all the samples of two groups by the software as is illustrated above (Figure 1A). The lincRNAs and intronic lncRNAs accounted for the highest proportion of the sequence reads of all the lncRNAs (Figure 1B). As shown in the boxplot, the discrete gene expression level distributions in a single sample (DOX1-3, BEC1-3) and the overall gene expression levels of two groups of samples were visually compared, we found that the gene expression of two groups was similar43 (Figure 1C). The mean gene expression in the samples is shown in Figure 1D, different groups of samples showed marked uniformity.
As lncRNA is less well conserved than mRNA, it was necessary to consider the conservation properties. Conservation scores of mRNA and lncRNA were determined by phastCons software. This showed that the lncRNA was less well-conserved than the mRNA (Figure 1E). The cumulative distribution of conservation scores and the conservation score probability density curve is presented in Figure 1F and G, respectively. The lncRNA of samples exhibited fair conservation.
Visualization of Differential Expression of lncRNAs in Cardiomyocytes
Before analyzing the difference between the DOX and BEC groups, Pearson’s Correlation Coefficient was adopted to compare the intra-group and inter-group differences (Figure 2A). We identified 301 lncRNAs that were differentially expressed between the DOX and BEC group. In total, 169 lncRNAs were elevated and 132 lncRNAs were downregulated in the BEC group compared with DOX group.
Hierarchical clustering analysis was performed on the differentially expressed lncRNAs. The differential expression of lncRNAs were presented by the volcano plot (Figure 2B) and heat map (Figure 2C). The green dots represent down-regulated lncRNAs, the red dots represent up-regulated lncRNAs, and the black dots represent non-differentially expressed lncRNAs. The top 10 elevated lncRNAs (ElncRNAs) in the BEC group are presented in Table 2.
 |
Table 2 The Top 10 ElncRNAs in the BEC Group |
Functional Annotation and Enrichment Analysis of ElncRNAs' Target Genes
To identify the regulatory functions and pathways associated with ElncRNAs and their targets, we performed GO and KEGG enrichment analyses. The GO enrichment analysis, including the biological process (BP), cellular component (CC), and molecular function (MF), of cis_target genes is displayed in Figure 3A–C. Both BP and MF results showed that cis_target genes mainly participated in transcription activities. In addition, they were enriched in inflammation-related biological process, such as the negative regulation of tumor necrosis factor production, negative regulation of macrophage activation, negative regulation of interleukin-1 beta production, and regulation of lymphocyte migration. CC outcomes showed that these genes may function in the nucleus.
On the other hand, the trans_targets of ElncRNAs were markedly enriched in cellular localization, in utero embryonic development, apoptotic processes, intracellular transport, chemotaxis, response to hypoxia, Golgi vesicle transport, and positive regulation of sodium ion transport (Figure S1A). CC analysis implied that trans_targets of ElncRNAs were chiefly located in the cytoplasm (Figure S1B). In terms of MF, trans_targets of ElncRNAs played significant roles in ATP binding, protein binding and homodimerization activity, zinc ion binding and superoxide dismutase activity (Figure S1C).
Human papillomavirus infection, hepatocellular and gastric carcinomas, Alzheimer’s diseases, the Wnt signaling pathway, mTOR signaling pathway, signaling pathways regulating stem cell pluripotency, and the Hippo signaling pathway were found to be markedly enriched in the KEGG pathways of cis_targets (Figure 4A). According to the novel classification of enrichment analysis, cis_targets were highly enriched in endocytosis and apoptosis pathways in cellular processes, the Wnt and mTOR signaling pathways in environmental information processing, protein processing in the endoplasmic reticulum in genetic information pathways, cancer pathways and viral carcinogenesis in human disease pathways, drug metabolism by cytochrome P450 and other enzymes in metabolism pathways, and olfactory transduction and adrenergic signaling in cardiomyocytes in the organismal systems pathways (Figure 4B).
In terms of the KEGG pathways, trans_targets of ElncRNAs were principally involved in pathways related to cancer, the PI3K-Akt signaling pathway, focal adhesion pathway, apoptosis pathway, cell cycle pathway, TNF signaling pathway, p53 signaling pathway, and fatty acid metabolism pathway (Figure S2A). We concluded that, based on the novel classification, the dominant pathways in which trans_targets were involved were mostly responsible for endocytosis and regulation of the actin cytoskeleton in cellular processes, the PI3K-Akt signaling pathway, and cytokine-cytokine receptor interaction in environmental information processing, RNA transport in genetic information processing, pathways in cancer in human diseases, and olfactory transduction and chemokine signaling in the organismal system pathways (Figure S2B).
Crucial Signaling Pathways in DIC
Based on the analyses above, we can conclude that inflammation or senescence play a pivotal role in DIC. The inflammation and senescence associated signaling pathways and networks are presented in Figure 5A and Figure 5B, respectively. The PI3K-Akt pathway and mTOR pathway were found to be markedly involved in the inflammation and senescence pathways (Figure 5C). Intriguingly, these are in line with the GO and KEGG enrichment analyses. From the Figure 5A–C, p53 is found not only central to DIC-related pathways, senescence and apoptosis, but also key to exosome-mediated secretion, implying its significant role in DIC (Figure 5D). Moreover, it is known that inflammation interacts with senescence. On the one hand, inflammation is a crucial factor for senescence and can accelerate it.44,45 On the other hand, senescence can induce senescence-associated inflammatory response (SIR), which is dependent on p53 activity.46 Actually, senescence is a consequence of inflammation response, which is more dominant than senescence in DIC.
 |
Figure 5 The crucial signaling pathways. (A) The chemokine signaling pathway. (B) The cellular senescence signaling pathway. (C) The mTOR signaling pathway. (D) The p53 signaling pathway. |
The Verification Experiments in vitro
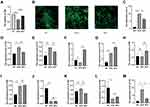
The above analyses suggested that injured cardiomyocytes can take up BMSC-Exos, which suppress the inflammatory response or oxidative stress-induced cell death caused by DOX. The ElncRNAs from exosomes may contribute to these rescue effects. To verify this hypothesis and our sequencing results, we performed a CCK-8 assay and found that cardiomyocytes senescence or death induced by DOX was reversed by BMSC-Exos (Figure 6A). To verify BMSC-Exos can downregulate the level of oxidative levels, we also detected the ROS levels in the NC, DOX, and BEC group. The ROS level was increased in the DOX group and was reduced in the BEC group (Figure 6B). And the qRT-PCR results show that cellular inflammation was inhibited in the BEC group (Figure 6C).
To verify whether exosomal lncRNAs were upregulated in the BEC group and whether the findings were plausible, we selected the top-10 elevated lncRNAs in the sequence results for verification, including MSTRG.75967.15, MSTRG.51708.15, MSTRG.98097.4, ENSMUST00000162380, MSTRG.94952.19, MSTRG.4585.1, MSTRG.79460.7, MSTRG.38414.10, MSTRG.58791.2 and MSTRG.12633.8. To this end, we extracted the total RNAs from the NC group, DOX group, and BEC group of cells, and conducted qRT-PCR experiments for these genes (Figure 6D–M). Of these lncRNAs, MSTRG.98097.4 and MSTRG.58791.2 were found to be reduced in the DOX group and increased in the BEC group. The decrease in the DOX group may indicate that these ElncRNAs are protective regulators against DOX-mediated injury, while the increase in the BEC group may indicate that they are released by exosomes. Thus, we identified them as effector molecules in BMSC-Exos.
The lncRNA-mRNA Network of Candidate Regulatory lncRNAs and Core Genes Mitigating DIC
As is analyzed in Figure 5, p53 has played a vital role during the pathophysiological processes of DIC. To explore other crucial genes in DIC, we further identified the target genes of candidate lncRNAs, MSTRG.98097.4 and MSTRG.58791.2, and drew a venn diagram (Figure 7A). Followingly, we constructed the lncRNA-mRNA network and exhibited the core target genes (Figure 7B and Table 3).
 |
Table 3 The Target Genes of MSTRG.58791.2 and MSTRG.98097.4 |
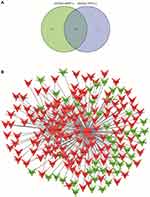 |
Figure 7 The lncRNA-mRNA network between two candidate lncRNAs and their target genes. (A) The common target genes of MSTRG.58791.2 and MSTRG.98097.4. (B) The network and core genes involving in DIC. |
The Verification Experiments in vivo
The exosome is cup-shaped in the photography (Figure 8A). The diameter distribution of exosome is about 100 nm (Figure 8B). The treatment of exosomes is illustrated in Figure 8C. The survival rate of DOX+BMSC-Exos increased (Figure 8D). And the fibrosis of cardiomyocytes decreased (Figure 8E) in the DOX+BMSC-Exo group compared with DOX group. The results of qRT-PCR showed that inflammation-related mRNAs (IL-1β, IL-6) were repressed in the BMSC-Exo group (Figure 8F and G). And the expression of MSTRG.58791.2 decreased in the heart tissue in the DOX group and increased in the DOX+BMSC-Exo group. While the expression of MSTRG.98097.4 is contrary to the MSTRG.58791.2 (Figure 8H and I). Finally, we summarized the process of our study in Figure 9.
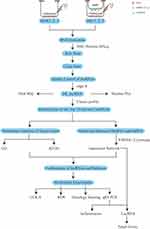 |
Figure 9 The whole process of the study. |
Discussion
At present, cardiovascular disease and cancer are the top two killers threatening human health. The side effects of DOX on the heart are called doxorubicin-induced cardiotoxicity (DIC). Clinical manifestations of cardiotoxicity, such as arrhythmia, myocarditis, and heart failure, greatly limit the clinical use of DOX.
Currently, the only recognized drug that prevent doxorubicin-induced cardiotoxicity (DIC) is dexrazoxane (DZR), but its effect is limited and there is a risk of secondary tumor or bone marrow suppression of DZR. It is vital to elucidate the pathogenesis of DIC and find effective therapeutic targets for the prevention and treatment of DIC.
Studies have shown that reactive oxygen species (ROS) or oxidative stress plays a central role in the emergence of DIC, which causes damage to cells and release cytokines that activate inflammation response.47 Except nanoparticles’ anti-inflammatory and therapeutic characteristic,48,49 the therapeutic role of mesenchymal stem cells derived exosomes on the heart has been confirmed.50 In this study, we compared the differential expression of lncRNAs between the DOX and BEC group and identified 169 lncRNAs were elevated (ElncRNAs) in the BEC group compared with the DOX group. The target genes of ElncRNAs were predicted by two different methods based on the positional relationship and expressive relationship, obtaining cis_targets and trans_targets, respectively.
The GO analysis of cis_targets displayed that they were particularly correlated with negative regulation of inflammation, such as negative regulation of tumor necrosis factor production, negative regulation of macrophage activation, negative regulation of interleukin-1 beta production, regulation of lymphocyte of migration and negative regulation of nitric-oxide synthase, suggesting that oxidative stress and inflammation play pivotal roles in DIC. The results were also in line with the previous studies.51–53
The KEGG analysis of cis_targets revealed that the Wnt pathway, mTOR pathway, and Hippo pathway were involved in the regulation of DIC. The Wnt pathway was reported to be involved in the process of arrhythmic cardiomyopathy,54 cell cycle and proliferation.55 The mTOR pathway has been reported to be a key pathway in DOX-induced cardiomyopathy.4 The Hippo pathway can modify physiological processes of cardiomyocytes, including development, proliferation, and signal transmission, and can be targeted to promote regeneration of cardiomyocytes.56,57 These pathways may be explored for regulating DIC.
While trans_targets were primarily enriched in the biological process (BP) of apoptosis, intracellular transport and Golgi vesicle transport. The Golgi complex assists the late sorting endosome to form the multivesicular body of exosomes, which are then released from the host cells into extracellular fluid through exocytosis and fuse with recipient cells, releasing their cargoes.11 The results of BP supported the hypothesis that trans_targets may participate in the biogenesis of exosomes, which executed the therapeutic functions for DIC. The pathways identified in the aspect of molecular function (MF) of GO show that trans_targets mainly participate in the activities of ATP binding and enzyme binding, which may supply the energy for exosomes biogenesis.
The KEGG analysis of trans_targets indicated that they mainly take part in pathophysiological activities of cancer, cell cycle, apoptosis, and fatty acid metabolism. It is known that oxidative stress can induce inflammation and apoptosis. Also, fatty acid metabolism is closely related to inflammation, its dysfunction can induce ferroptosis,58,59 which is iron-dependent and likely to occur in DIC.5 The enrichment of fatty acid metabolism pathways may be related to the regulation of ferroptosis by exosomal lncRNAs. Furthermore, trans_targets were also shown to enrich in the PI3K-Akt signaling pathway, TNF signaling pathway, and p53 signaling pathway. PI3K-Akt is not only associated with inflammation (Figure 6A), but also is related to the regeneration of cardiomyocytes, along with the Wnt and Hippo signaling pathways.57 In addition, PI3K-Akt, mTOR, and p53 pathways construct to establish a complex network of cell senescence (Figure 6B). P53 is a tumor suppressor protein that also plays a role in programmed cell death, including apoptosis, senescence, and ferroptosis, and is key to the regulation of the PI3K-Akt pathway and Wnt pathway (Figure 6A‒D). The PPI network provide a whole picture of interaction between candidate lncRNA-mRNA. The common target genes facilitated us to identify the key genes or pathways.
Combining the results of enrichment analyses of cis_targets and trans_targets, we deduced that exosomes secreted by BMSCs are likely to rescue the DOX-injured cardiomyocytes by releasing lncRNAs. Our verification assays showed that exosomes derived from BMSC potently increased the viability, reduced oxidative stress and inhibited the inflammation response in DOX-injured cardiomyocytes, supporting previous reports,60,61 and qRT-PCR results substantiated that MSTRG.98097.4 and MSTRG.58791.2 down-regulated in the DOX group and up-regulated in the BEC group. But in animal experiments, only the expression of MSTRG.58791.2 was consistent with the cell experiment, the expression of MSTRG.98097.4 is contrary to the cell experiment. The reason may be that both myocardial fibroblasts and cardiomyocytes consist of the heart. When we extracted RNA from the heart tissues, it is unavoidable that the RNA of fibroblasts interfers with the results. This interference may be more obvious for MSTRG.98097.4. Although the result of MSTRG.98097.4 is contrary to the cell experiment, but it is not meaningless, which is also shown that exosomes execute the therapeutic role for DIC. The pathway may be via mediating lncRNAs in the cardiomyocytes instead of releasing by exogenous exosomes.
There are some limitations in this study. Firstly, the sequencing method is indirect, and we cannot be sure that the ElncRNAs are exactly derived from exogenous exosomes although there are some evidences pointing that the ElncRNAs are from the BMSC-Exos. Future work should be concentrated on labeling the candidate lncRNAs in exosomes from host cells to visualize their movement. Additionally, it is crucial to perform loss-and-gain experiments to verify the function of ElncRNAs.
Conclusion
Bone marrow mesenchymal stem cell-derived exosomes inhibit the inflammatory response of cardiomyocytes by mediating lncRNA, thereby alleviating DIC. In addition, a kind of lncRNA, MSTRG.58791.2, which may be derived from exosomes and regulate DIC, was preliminarily screened out in this study for further study.
Abbreviations
BEC, BMSC-derived exosomes coculture; BMSC, bone marrow stem cells; BMSC-Exos, bone marrow mesenchymal stem cells-derived exosomes; BP, biological process; CC, cellular component; DIC, doxorubicin-induced cardiotoxicity; DOX, doxorubicin; DZR, dexrazoxane; ElncRNA, elevated lncRNAs; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; lncRNA, long non-coding RNAs; MF, molecular function; SIR, senescence-associated inflammatory response.
Data Sharing Statement
The data and materials of this study are included within the article.
Ethics Committee Approval
The animal experiments were approved by the Ethics Committee of Qingdao University.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by The National Natural Science Foundation of China (no. 82172574), the National Natural Science Foundation of China (no. 81871231), The Natural Science Foundation of Shandong Province (no. ZR2020MH016), Research Planning Project of Shandong Higher Medical Education Research Center (no. YJKT202171), Project of China Association of Chinese Medicine (no. 2021HH-006), Shandong Chinese Medicine Science, Technology Project (no. 2021M164) and Youth Innovation and Science and Technology Plan of Colleges and Universities in Shandong Province (no. 2019KJK016).
Disclosure
The authors have declared that no competing interest exists.
References
1. Maejima Y, Adachi S, Ito H, Hirao K, Isobe M. Induction of premature senescence in cardiomyocytes by doxorubicin as a novel mechanism of myocardial damage. Aging Cell. 2008;7(2):125–136. doi:10.1111/j.1474-9726.2007.00358.x
2. Arola OJ, Saraste A, Pulkki K, Kallajoki M, Parvinen M, Voipio-Pulkki LM. Acute doxorubicin cardiotoxicity involves cardiomyocyte apoptosis. Cancer Res. 2000;60(7):1789–1792.
3. Wang Y, Gao W, Shi X, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547(7661):99–103. doi:10.1038/nature22393
4. Bartlett JJ, Trivedi PC, Pulinilkunnil T. Autophagic dysregulation in doxorubicin cardiomyopathy. J Mol Cell Cardiol. 2017;104:1–8. doi:10.1016/j.yjmcc.2017.01.007
5. Fang X, Wang H, Han D, et al. Ferroptosis as a target for protection against cardiomyopathy. Proc Natl Acad Sci U S A. 2019;116(7):2672–2680. doi:10.1073/pnas.1821022116
6. Räsänen M, Degerman J, Nissinen TA, et al. VEGF-B gene therapy inhibits doxorubicin-induced cardiotoxicity by endothelial protection. Proc Natl Acad Sci USA. 2016;113(46):13144–13149. doi:10.1073/pnas.1616168113
7. Brown SA, Sandhu N, Herrmann J. Systems biology approaches to adverse drug effects: the example of cardio-oncology. Nat Rev Clin Oncol. 2015;12(12):718–731. doi:10.1038/nrclinonc.2015.168
8. Curigliano G, Cardinale D, Dent S, et al. Cardiotoxicity of anticancer treatments: epidemiology, detection, and management. CA Cancer J Clin. 2016;66(4):309–325. doi:10.3322/caac.21341
9. Gutteridge JM. Lipid peroxidation and possible hydroxyl radical formation stimulated by the self-reduction of a doxorubicin-iron (III) complex. Biochem Pharmacol. 1984;33(11):1725–1728. doi:10.1016/0006-2952(84)90340-X
10. Han X, Zhou Y, Liu W. Precision cardio-oncology: understanding the cardiotoxicity of cancer therapy. NPJ Precis Oncol. 2017;1(1):31. doi:10.1038/s41698-017-0034-x
11. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:6478. doi:10.1126/science.aau6977
12. Dai J, Su Y, Zhong S, et al. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct Target Ther. 2020;5(1):145. doi:10.1038/s41392-020-00261-0
13. Zong T, Yang Y, Lin X, et al. 5’-tiRNA-Cys-GCA regulates VSMC proliferation and phenotypic transition by targeting STAT4 in aortic dissection. Mol Ther Nucleic Acids. 2021;26:295–306. doi:10.1016/j.omtn.2021.07.013
14. Zou Y, Yang Y, Fu X, et al. The regulatory roles of aminoacyl-tRNA synthetase in cardiovascular disease. Mol Ther Nucleic Acids. 2021;25:372–387. doi:10.1016/j.omtn.2021.06.003
15. Fu X, He X, Yang Y, et al. Identification of transfer RNA-derived fragments and their potential roles in aortic dissection. Genomics. 2021;113(5):3039–3049. doi:10.1016/j.ygeno.2021.06.039
16. Li X, Yang Y, Wang Z, et al. Targeting non-coding RNAs in unstable atherosclerotic plaques: mechanism, regulation, possibilities, and limitations. Int J Biol Sci. 2021;17(13):3413–3427. doi:10.7150/ijbs.62506
17. Li M, Yang Y, Wang Z, et al. Piwi-interacting RNAs (piRNAs) as potential biomarkers and therapeutic targets for cardiovascular diseases. Angiogenesis. 2021;24(1):19–34. doi:10.1007/s10456-020-09750-w
18. Zhang YF, Xu HM, Yu F, et al. Crosstalk between microRNAs and peroxisome proliferator-activated receptors and their emerging regulatory roles in cardiovascular pathophysiology. PPAR Res. 2018;2018:8530371. doi:10.1155/2018/8530371
19. Li M, Yang Y, Zong J, et al. miR-564: a potential regulator of vascular smooth muscle cells and therapeutic target for aortic dissection. J Mol Cell Cardiol. 2022;170:100–114. doi:10.1016/j.yjmcc.2022.06.003
20. Zhang Y, Jia DD, Zhang YF, et al. The emerging function and clinical significance of circRNAs in thyroid cancer and autoimmune thyroid diseases. Int J Biol Sci. 2021;17(7):1731–1741. doi:10.7150/ijbs.55381
21. Tian C, Yang Y, Bai B, et al. Potential of exosomes as diagnostic biomarkers and therapeutic carriers for doxorubicin-induced cardiotoxicity. Int J Biol Sci. 2021;17(5):1328–1338. doi:10.7150/ijbs.58786
22. Tavakoli Dargani Z, Singla DK. Embryonic stem cell-derived exosomes inhibit doxorubicin-induced TLR4-NLRP3-mediated cell death-pyroptosis. Am J Physiol Heart Circ Physiol. 2019;317(2):H460–h471. doi:10.1152/ajpheart.00056.2019
23. Singla DK, Johnson TA, Tavakoli Dargani Z. Exosome Treatment enhances anti-inflammatory M2 macrophages and reduces inflammation-induced pyroptosis in doxorubicin-induced cardiomyopathy. Cells. 2019;8(10):1224. doi:10.3390/cells8101224
24. Yang Y, Li M, Liu Y, et al. The lncRNA punisher regulates apoptosis and mitochondrial homeostasis of vascular smooth muscle cells via targeting miR-664a-5p and OPA1. Oxid Med Cell Longev. 2022;2022:5477024. doi:10.1155/2022/5477024
25. Ponnusamy M, Liu F, Zhang YH, et al. Long noncoding RNA CPR (Cardiomyocyte Proliferation Regulator) regulates cardiomyocyte proliferation and cardiac repair. Circulation. 2019;139(23):2668–2684. doi:10.1161/CIRCULATIONAHA.118.035832
26. Cai B, Ma W, Ding F, et al. The long noncoding RNA CAREL controls cardiac regeneration. J Am Coll Cardiol. 2018;72(5):534–550. doi:10.1016/j.jacc.2018.04.085
27. Liu N, Kataoka M, Wang Y, et al. LncRNA LncHrt preserves cardiac metabolic homeostasis and heart function by modulating the LKB1-AMPK signaling pathway. Basic Res Cardiol. 2021;116(1):48. doi:10.1007/s00395-021-00887-3
28. Sun L, Zhu W, Zhao P, et al. Long noncoding RNA UCA1 from hypoxia-conditioned hMSC-derived exosomes: a novel molecular target for cardioprotection through miR-873-5p/XIAP axis. Cell Death Dis. 2020;11(8):696. doi:10.1038/s41419-020-02783-5
29. Li KS, Bai Y, Li J, et al. LncRNA HCP5 in hBMSC-derived exosomes alleviates myocardial ischemia reperfusion injury by sponging miR-497 to activate IGF1/PI3K/AKT pathway. Int J Cardiol. 2021;342:72–81. doi:10.1016/j.ijcard.2021.07.042
30. Mao Q, Liang XL, Zhang CL, Pang YH, Lu YX. LncRNA KLF3-AS1 in human mesenchymal stem cell-derived exosomes ameliorates pyroptosis of cardiomyocytes and myocardial infarction through miR-138-5p/Sirt1 axis. Stem Cell Res Ther. 2019;10(1):393. doi:10.1186/s13287-019-1522-4
31. Dubey RK, Gillespie DG, Mi Z, Jackson EK. Exogenous and endogenous adenosine inhibits fetal calf serum-induced growth of rat cardiac fibroblasts: role of A2B receptors. Circulation. 1997;96(8):2656–2666. doi:10.1161/01.CIR.96.8.2656
32. Li RK, Mickle DA, Weisel RD, Zhang J, Mohabeer MK. In vivo survival and function of transplanted rat cardiomyocytes. Circ Res. 1996;78(2):283–288. doi:10.1161/01.RES.78.2.283
33. Zhu J, Liu B, Wang Z, et al. Exosomes from nicotine-stimulated macrophages accelerate atherosclerosis through miR-21-3p/PTEN-mediated VSMC migration and proliferation. Theranostics. 2019;9(23):6901–6919. doi:10.7150/thno.37357
34. Kong L, Zhang Y, Ye ZQ, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35(Web Server issue):W345–349. doi:10.1093/nar/gkm391
35. Sun L, Luo H, Bu D, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41(17):e166. doi:10.1093/nar/gkt646
36. Finn RD, Bateman A, Clements J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Databaseissue):D222–230. doi:10.1093/nar/gkt1223
37. Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Cpat: LW. Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41(6):e74. doi:10.1093/nar/gkt006
38. Hubisz MJ, Pollard KS, Siepel A. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief Bioinform. 2011;12(1):41–51. doi:10.1093/bib/bbq072
39. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
40. Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(7):D808–815. doi:10.1093/nar/gks1094
41. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
42. Gao L, Wang L, Dai T, et al. Tumor-derived exosomes antagonize innate antiviral immunity. Nat Immunol. 2018;19(3):233–245. doi:10.1038/s41590-017-0043-5
43. Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi:10.1038/nbt.1621
44. Jurk D, Wilson C, Passos JF, et al. Chronic inflammation induces telomere dysfunction and accelerates ageing in mice. Nat Commun. 2014;5(1):4172. doi:10.1038/ncomms5172
45. Desdín-Micó G, Soto-Heredero G, Aranda JF, et al. T cells with dysfunctional mitochondria induce multimorbidity and premature senescence. Science. 2020;368(6497):1371–1376. doi:10.1126/science.aax0860
46. Pribluda A, Elyada E, Wiener Z, et al. A senescence-inflammatory switch from cancer-inhibitory to cancer-promoting mechanism. Cancer Cell. 2013;24(2):242–256. doi:10.1016/j.ccr.2013.06.005
47. Quagliariello V, De Laurentiis M, Rea D, et al. The SGLT-2 inhibitor empagliflozin improves myocardial strain, reduces cardiac fibrosis and pro-inflammatory cytokines in non-diabetic mice treated with doxorubicin. Cardiovasc Diabetol. 2021;20(1):150. doi:10.1186/s12933-021-01346-y
48. Li X, Yang Y, Wang Z, et al. Multistage-responsive nanocomplexes attenuate ulcerative colitis by improving the accumulation and distribution of oral nucleic acid drugs in the colon. ACS Appl Mater Interfaces. 2022;14(1):2058–2070. doi:10.1021/acsami.1c21595
49. Qi HZ, Yang J, Yu J, et al. Glucose-responsive nanogels efficiently maintain the stability and activity of therapeutic enzymes. Nanotechnol Rev. 2022;11(1):1511–1524. doi:10.1515/ntrev-2022-0095
50. Zhao J, Li X, Hu J, et al. Mesenchymal stromal cell-derived exosomes attenuate myocardial ischaemia-reperfusion injury through miR-182-regulated macrophage polarization. Cardiovasc Res. 2019;115(7):1205–1216. doi:10.1093/cvr/cvz040
51. Ni C, Ma P, Wang R, et al. Doxorubicin-induced cardiotoxicity involves IFNγ-mediated metabolic reprogramming in cardiomyocytes. J Pathol. 2019;247(3):320–332. doi:10.1002/path.5192
52. Mu H, Liu H, Zhang J, et al. Ursolic acid prevents doxorubicin-induced cardiac toxicity in mice through eNOS activation and inhibition of eNOS uncoupling. J Cell Mol Med. 2019;23(3):2174–2183. doi:10.1111/jcmm.14130
53. Xia N, Bollinger L, Steinkamp-Fenske K, Förstermann U, Li H. Prunella vulgaris L. Upregulates eNOS expression in human endothelial cells. Am J Chin Med. 2010;38(3):599–611. doi:10.1142/S0192415X10008081
54. Austin KM, Trembley MA, Chandler SF, et al. Molecular mechanisms of arrhythmogenic cardiomyopathy. Nat Rev Cardiol. 2019;16(9):519–537. doi:10.1038/s41569-019-0200-7
55. Buikema JW, Mady AS, Mittal NV, et al. Wnt/β-catenin signaling directs the regional expansion of first and second heart field-derived ventricular cardiomyocytes. Development. 2013;140(20):4165–4176. doi:10.1242/dev.099325
56. Wang J, Liu S, Heallen T, Martin JF. The Hippo pathway in the heart: pivotal roles in development, disease, and regeneration. Nat Rev Cardiol. 2018;15(11):672–684. doi:10.1038/s41569-018-0063-3
57. Lin Z, Zhou P, von Gise A, et al. Pi3kcb links Hippo-YAP and PI3K-AKT signaling pathways to promote cardiomyocyte proliferation and survival. Circ Res. 2015;116(1):35–45. doi:10.1161/CIRCRESAHA.115.304457
58. Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi:10.1016/j.cell.2012.03.042
59. Stockwell BR, Friedmann Angeli JP, Bayir H, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi:10.1016/j.cell.2017.09.021
60. Milano G, Biemmi V, Lazzarini E, et al. Intravenous administration of cardiac progenitor cell-derived exosomes protects against doxorubicin/trastuzumab-induced cardiac toxicity. Cardiovasc Res. 2020;116(2):383–392. doi:10.1093/cvr/cvz108
61. Xia W, Chen H, Xie C, Hou M. Long-noncoding RNA MALAT1 sponges microRNA-92a-3p to inhibit doxorubicin-induced cardiac senescence by targeting ATG4a. Aging. 2020;12(9):8241–8260. doi:10.18632/aging.103136
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.