Back to Journals » OncoTargets and Therapy » Volume 8
Downregulation of microRNA-132 by DNA hypermethylation is associated with cell invasion in colorectal cancer
Authors Qin J, Ke J, Xu J , Wang F, Zhou Y, Jiang Y, Wang Z
Received 1 July 2015
Accepted for publication 13 October 2015
Published 7 December 2015 Volume 2015:8 Pages 3639—3648
DOI https://doi.org/10.2147/OTT.S91560
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Faris Farassati
Jun Qin, Jing Ke, Junfei Xu, Feiran Wang, Youlang Zhou, Yasu Jiang, Zhiwei Wang
Department of General Surgery, Affiliated Hospital of Nantong University, Nantong, Jiangsu Province, People’s Republic of China
Abstract: microRNAs (miRNAs) are small, noncoding RNAs that are involved in many biological processes, and aberrant regulation of miRNAs is always associated with cancer progression and development. Abnormal expression of miRNA-132 (miR-132) has been found in some types of cancer, but the effects and potential mechanisms of miR-132 in colorectal cancer (CRC) have not been explored to date. In this study, quantitative real-time polymerase chain reaction was used to investigate the level of miR-132 in CRC tissues and their paired adjacent normal tissues. Bioinformatics analysis indicated that the mechanism underlying the tumor suppressor role of miR-132 in CRC cells may play a role in tumor suppression by targeting paxillin. Furthermore, methylation-specific polymerase chain reaction was performed to evaluate the methylation status of the miR-132 regulatory region. A DNA methyltransferase inhibitor, 5-aza-2'-deoxycytidine, was used to activate the expression of miR-132 in CRC cells in vitro. Downregulation of miR-132 may occur as a result of hypermethylation and implies a poor prognosis in CRC; therefore, triggering miR-132 reexpression by using DNA methyltransferase inhibitors may be a potential molecular therapeutic target for CRC.
Keywords: microRNA-132, colorectal cancer, DNA methylation, invasion
Introduction
Worldwide, colorectal cancer (CRC) is the second and third most common cancer type in women and men, respectively, and shows a rapid progression of the tumor.1 In economically developing countries, especially in People’s Republic of China, CRC is becoming more prevalent.2 Substantial progress in the diagnosis and treatment of CRC has been achieved, but the overall 5-year survival rate of CRC remains low.3 Although tumor migration and metastasis are known to be the key causes of death in patients with CRC, the molecular pathogenesis of this disease remains largely unclear. Therefore, the identification of biomarkers for CRC would be helpful to predict the metastatic progression and prognosis and to provide a basis for targeted therapy schemes.
microRNAs (miRNAs) represent a large family of endogenously expressed, noncoding RNAs with a length of 18–25 nucleotides, which regulate gene expression by inducing RNA degradation or interfering with translation. Some miRNAs have been shown to be involved in various biological processes such as inflammation, tissue development, cell survival, proliferation, and metabolism by targeting multiple protein-coding genes through partial base pairing to the 3′-untranslated region (UTR) of the target gene.4–6 Aberrant miRNA expression has been reported in a variety of human diseases, particularly cancer. In this study, we show that miRNA-132 (miR-132), transcribed from an intergenic region on human chromosome 17, is aberrantly expressed in lung and pancreatic cancers and in the endothelium of breast carcinoma tumors.7,8 A recent report indicated that miR-132 was significantly downregulated in CRC tissues with distant metastases, and the ectopic expression of miR-132 markedly inhibited cell invasion and epithelial–mesenchymal transition in CRC cell lines by targeting zinc finger E-box binding homeobox 2.9 Furthermore, the previous study has reported that silencing of miR-132 by promoter CpG island methylation may be an important mechanism in the carcinogenesis of both prostate and pancreatic cancers.10,11 However, the molecular mechanisms underlying miR-132 epigenetic regulation in CRC carcinogenesis have not yet been clarified.
DNA methylation is the most widely studied epigenetic event, especially in CpG islands, and leads to transcriptional gene silencing. DNA methylation plays an important role in tumorigenesis. CpG island methylation of tumor suppressor genes, resulting in the inactivation of gene transcription, has become an important subject for cancer epigenetics research.12,13 Numerous tumor suppressor genes inactivated by promoter CpG island methylation have been found in a variety of cancer tissues and cells, especially in CRC.
The aim of this study was to investigate whether the downregulation of miR-132 originates from hypermethylation of the genomic region upstream of miR-132 in CRC. Therefore, we analyzed the DNA methylation of CpG islands in the promoter region of miR-132 in patients with CRC and found that the downregulation of miR-132 in CRC tissues could result from hypermethylation of promoter CpG islands.
Materials and methods
Patients and tissue samples
Surgical specimens of cancer tissues and adjacent nontumor tissue samples were obtained from 36 patients with CRC who underwent surgery at the Affiliated Hospital of Nantong University between 2013 and 2014. None of the patients had received chemotherapy or radiotherapy before surgery excision. After collection, all tissue samples were immediately frozen in liquid nitrogen and stored at −80°C until use. Tumor, node, and metastasis stage was classified according to the International Union Against Cancer.
Written informed consent was obtained from each patient, and research protocols were approved by the Institutional Ethics Board of the Affiliated Hospital of Nantong University.
Cell culture and transfection
The human CRC cell lines HCT116, SW480, HT-29, HCT-8, SW620, and LoVo and the normal colon epithelium cell line (fetal human cells [FHC]) were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, People’s Republic of China) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) or Roswell Park Memorial Institute-1640 medium supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA). Cells were cultured at 37°C in a humidified incubator containing 5% CO2. Cells were transfected using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s guidelines.
Quantitative real-time polymerase chain reaction
Total RNA was extracted from the cells and tissues with TRIzol reagent (Thermo Fisher Scientific) and reversely transcribed to cDNA using a stem-loop reverse transcription primer for miRNA detection. The reverse transcription of miR-132 and internal control U6 was performed using reverse transcriptase Moloney murine leukemia virus (TaKaRa, Tokyo, Japan). Quantitative real-time polymerase chain reaction (PCR) was performed using SYBR Premix ex Taq (TaKaRa) following the manufacturer’s protocol and using a preheated real-time instrument (Thermo Fisher Scientific). Three independent experiments were conducted to analyze the relative gene expression, and each sample was tested in triplicate. All the mRNA quantification data were normalized by the Ct value of U6 (internal control) using a 2−ΔΔCt relative quantification method.
Luciferase activity assay
A miR-132 expression plasmid (pcDNA6.2-miR-132) was constructed with synthetic miR-132 oligonucleotides and the pcDNA6.2-GW/EmGFP vector. A negative control (NC; mock miRNA) vector was constructed in the same way as the abovementioned pcDNA6.2-miR-132 plasmids, except that the inserted miRNA oligomer was synthesized according to the random genetic sequences. The 3′-UTR of paxillin mRNA and a mutant variant were generated by PCR fragments and cloned into the XbaI site of a pGL3-basic vector (Promega Corporation, Fitchburg, WI, USA) and termed paxillin-wt-3′-UTR and paxillin-mt-3′-UTR, respectively. All constructs were confirmed by DNA sequence analysis. For luciferase assay, SW480 cells were seeded in six-well plates the day before transfection and were transfected with the pGL3 reporter plasmid (250 ng/well), pRL-TK luciferase reporter plasmid (25 ng/well), miR-132 mimics, and an NC, using Lipofectamine 3000. Luciferase activities were measured at 24 hours after transfection with a Dual Luciferase Reporter Assay Kit (Promega Corporation) according to the manufacturer’s protocol. Chemically synthesized RNAs, including NC, miR-132 mimics, and a miR-132 inhibitor, were obtained from Gene Pharma (Shanghai, People’s Republic of China).
Treatment with 5-aza-2′-deoxycytidine
SW480 and LoVo cells were seeded at a density of 5×106/well in six-well plates, cultured for 24 hours, and then treated with 10 μM of the DNA demethylating agent 5-aza-2′-deoxycytidine (5-Aza-CdR; Sigma-Aldrich Co., St Louis, MO, USA) for 3 days, during which the drug-containing medium was replaced daily.
Methylation-specific PCR analysis
Methylation-specific PCR (MSP) analysis was used to analyze the methylation of miR-132 upstream region in cell lines and tissues. Genomic DNA extracted from cell lines and tissues was modified with bisulfite reagents following the manufacturer’s instructions (Zymo Research, Irvine, CA, USA). The bisulfite-modified DNA was purified using a QIAquick Gel Extraction Kit (Qiagen NV, Venlo, the Netherlands) according to the manufacturer’s instructions. PCR amplification was carried out using 20 ng of bisulfite-modified DNA. The MSP primers and PCR conditions were as follows: methylated: forward 5′-TTTTTTGGGATATTTTTGACGTTAC-3′ and reverse 5′-CCGACTAAAAACTCTACTACTCCG-3′, with PCR settings of 95°C for 10 minutes, followed by 35 cycles of 96°C for 39 seconds, 67°C for 40 seconds, and 72°C for 40 seconds, with a final elongation step at 72°C for 10 minutes; unmethylated: forward 5′-TTTTTGGGATATTTTTGATGTTATG-3′ and reverse 5′-CCAACTAAAAACTCTACTACTCCAC-3′, with PCR settings of 95°C for 10 minutes, followed by 35 cycles of 96°C for 39 seconds, 58°C for 30 seconds, and 72°C for 30 seconds, with a final elongation step at 72°C for 10 minutes.
Bisulfite-assisted PCR assay
Methylation statuses of the CpG sites in a miR-132-associated CpG island were determined by bisulfite-assisted PCR sequencing. In brief, DNAs from CRC tissues and adjacent nontumor tissues were bisulfite converted as described earlier. Bisulfite-modified DNA was amplified by methylation-unbiased primers designed using the MethPrimer program (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi). The bisulfite-assisted PCR primers used were as follows: forward 5′-GGGTTATTTTGGGTTTTTTTATTTT-3′ and reverse 5′-CCCTCCTCAATTCCTAAACCTAA-3′, with initial denaturation for 2 minutes at 96°C, followed by 35 cycles for 10 seconds at 94°C, 5 seconds at 59°C, and 30 seconds at 72°C, with a final elongation for 10 minutes at 72°C. Amplification products (186 bp, from positions −1,717 to −1,532) were sequenced after cloning into the prokaryotic expression vector (Promega Corporation). Using blue/white screening, ten clones of each specimen were sequenced using M13 forward or reverse primers.
Transwell assay
The invasive capacity of the cells was determined using transwell chambers containing a polycarbonate membrane filter of 4-μm pore size and coated with Matrigel (BD Biosciences, San Jose, CA, USA). Briefly, SW480 and LoVo cells (1×105/well) were incubated with 5-Aza-CdR for 3 days, then they were separately detached from the tissue culture plates, washed, added to 100 μL of serum-free medium, and then placed in the upper chamber of the inserts. DMEM (Thermo Fisher Scientific) containing 10% fetal bovine serum (500 μL) was added to the lower chamber as the chemotactic factor. After culture with 5% CO2 at 37°C for 48 hours, the noninvasive cells on the upper surface were removed gently with a cotton swab, while the cells that had invaded to the lower side of the compartment were fixed and dyed with 0.1% crystal violet. Photographs of three randomly selected fields of the fixed cells were taken, and the numbers of invaded cells were counted. All the experiments were performed in triplicate.
Statistical analysis
Differences in mRNA and protein expression between tumor samples and the paired adjacent nontumor tissue samples were evaluated with paired-samples t-test, and P<0.05 was considered statistically significant. Statistical analyses were performed using the Statistical Package for the Social Sciences (Version 18.0; SPSS Inc., Chicago, IL, USA).
Results
miR-132 was significantly downregulated in CRC cell lines and CRC tissues
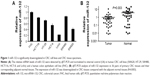
To assess the expression of miR-132 in CRC cell lines, we examined the expression of miR-132 in six human CRC cell lines (SW480, HCT116, HT-29, HCT-8, SW620, and LoVo) and a normal colon epithelium cell line (FHC). The expression level of miR-132 in FHC was significantly higher than that in any of the six CRC cell lines (SW480, HCT116, HT-29, HCT-8, SW620, or LoVo; Figure 1A). In addition, we found that the expression of miR-132 was highest in SW480 cells and lowest in LoVo cells, so we chose these two typical cell lines for further study.
Furthermore, quantitative real-time PCR analysis was conducted in 36 pairs of CRC tissues and matched adjacent normal tissue samples, and the results showed that the expression of miR-132 was downregulated in CRC tissues compared with adjacent normal tissues (P=0.003; Figure 1B).
These data indicate that the reduced expression of miR-132 might be involved in the development of CRC.
Paxillin is a novel target of miR-132 in CRC
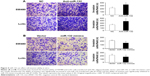
miRNAs perform biological functions by negatively regulating their target genes. In this study, targets of miR-132 in CRC were analyzed using the data predicted from the TargetScan database, PicTar, and miRanda (Figure 2A and B). To verify the direct effects of miR-132 on paxillin in CRC cells, we transfected SW480 and LoVo cells with a luciferase reporter plasmid containing the 3′UTR of paxillin mRNA. As shown in Figure 2C, miR-132 mimicked decreased the paxillin 3′UTR luciferase reporter activity in SW480 and LoVo cells. This effect was not observed when the nucleotides in the miR-132 binding site of the paxillin mRNA 3′UTR were mutated. Therefore, our data are consistent with the hypothesis that miR-132 represses paxillin protein levels via direct targeting of the 3′UTR of paxillin.
In addition, Western blot and transwell assay showed that, compared with cells transfected with the control vector, the overexpression of miR-132 resulted in decreased protein levels of paxillin and increased invasive ability in CRC cells, whereas paxillin expression was significantly increased in miR-132 inhibitor transfected cells. Similarly, cell-invasive ability was significantly enhanced after transfection with a miR-132 inhibitor (Figures 3 and 4).
In summary, these results demonstrated that miR-132 inhibited the expression of paxillin through posttranscriptional repression.
miR-132 is epigenetically silenced in CRC cell lines
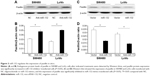
To study the regulation of miR-132 by DNA methylation, we searched the human genome database for the presence of CpG islands around miR-132 using CpG Island Searcher software (www.urogene.org/methprimer/) and identified a CpG island located from −1,714 to −54 bp upstream of miR-132 (Figure 5A).
To identify whether the low miR-132 expression in CRC was due to DNA methylation, we first examined the DNA methylation of miR-132 in CRC cell lines, thus we treated SW480 and LoVo cells with the DNA methylation inhibitor 5-Aza-CdR. The expression of miR-132 was upregulated in SW480 and LoVo cells when they were treated with 5-Aza-CdR, suggesting that the expression of miR-132 might be inhibited in these cells by DNA methylation (Figure 5B and C). Subsequently, we further examined the DNA methylation status using MSP in SW480 and LoVo cell lines. We found that the CpG islands were hypermethylated in all two CRC cell lines, which was consistent with the low expression of miR-132 in these CRC cell lines. When the CRC cells lines were treated with 5-Aza-CdR, the methylation level was decreased compared with the control group (Figure 5D).
Downregulation of miR-132 in CRC tissues due to the hypermethylation of CpG islands
We further analyzed the methylation status of the CpG islands in the CRC tissues and the adjacent non-tumor tissues of 36 randomly selected cases by using MSP. Representative MSP results of six cases are shown in Figure 5E. The methylation level in the 36 CRC tissues was higher than that in the adjacent nontumor tissues (P<0.05; Figure 5F), which was consistent with the low expression of miR-132 in the CRC tissues.
To further confirm the methylation status of CpG sites within the human miR-132 promoter, DNA sequencing was performed on PCR products of the 186-bp fragment (−1,717 to −1,532) obtained after the treatment of genomic DNA samples with sodium bisulfite. The 186-bp fragment was found to contain 12 CpG sites (Figure 6A, underlined). All samples from 20 paired CRC and adjacent nontumor tissues were successfully sequenced. The methylation frequencies of miR-132 promoter in CRC tissues and adjacent nontumor tissues were 32.18%±12.26% and 11.86%±5.71%, respectively. The methylation frequencies of miR-132 promoter in CRC tissues were higher than those in the adjacent nontumor tissues. Representative results of bisulfite sequencing analysis for gene promoters in CRC and adjacent nontumor tissue samples are shown in Figure 6B.
Ultimately, these findings provide powerful evidence that the expression of miR-132 in CRC is regulated by DNA methylation and that the downregulation of miR-132 in CRC is due to the hypermethylation of its CpG islands.
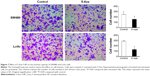
Effect of 5-Aza-CdR on invasive capacity of SW480 and LoVo cells
The invasive ability of cells treated with 5-Aza-CdR and of untreated cells (no 5-Aza-CdR) was evaluated by transwell assays. We found that the number of invading cells was significantly decreased in SW480 and LoVo cells treated with 5-Aza-CdR compared with the control group (P<0.05), indicating that the invasive capacity of 5-Aza-CdR-treated SW480 and LoVo cells was significantly decreased compared with the control group (Figure 7). Therefore, 5-Aza-CdR is able to inhibit the expression of paxillin by the upregulation of miR-132, thus inhibiting the invasive ability of CRC cells.
Discussion
There has been increasing interest in analyzing changes in the miRNA profile of various cancer cells and their roles in carcinogenesis.14 In a recent study,9 we used miRNA regulatory network analysis for tissue expression profiling of patients with CRC and identified that the levels of miR-132 were strongly decreased, in accordance with the role of miR-132 as a candidate tumor suppressor in a series of cancers, including hepatocellular carcinoma, prostate cancer, and ductal carcinoma in situ of the breast.10,15,16 In addition, a recent report showed that the dysregulation of miR-132 is associated with several neurological disorders, such as Parkinson’s disease, Alzheimer’s disease, schizophrenia, and tauopathies,17,18 suggesting a broader impact of this miRNA in diseases of brain development. Further research revealed that miR-132 exerts its tumor-suppressing effects through the inactivation of the Akt-signaling pathway, specifically by reducing Akt phosphorylation and the concentration of cyclin D1.11
Because the influence of cancerous miRNAs on cancer biology depends on the functions of the downstream targets they suppress, the targets of each miRNA need to be identified. A previous study reported that paxillin has been validated as a direct target of miR-137 in CRC, suggesting that the overexpression of paxillin induced by the suppression of miR-137 may promote tumor progression and metastasis and predict poor prognosis.19 In this study, our data showed that the overexpression of miR-132 significantly reduced the activity of a luciferase reporter containing the 3′-UTR sequence of paxillin, which has been confirmed as a target gene of miR-132.
Some studies have reported that the methylation of the promoter region of miR-132 in both prostate and pancreatic cancer samples is a major reason for the low expression of miR-132 in these tumors,10,11 but there is no evidence that the methylation of miR-132 in CRC is due to its low expression. Thus, in order to further clarify the mechanism of miR-132 in suppressing tumorigenesis in CRC, we investigated whether the downregulation of miR-132 in CRC tissues might be due to the hypermethylation of CpG sequences in its promoter. It is well known that carcinoma results from the combined forces of both genetic and epigenetic events. DNA methylation is the main epigenetic event in humans, and the changes in DNA methylation pattern play an important role in tumorigenesis.20 DNA methylation, especially in CpG islands, leads to transcriptional gene silencing. To date, the inactivation of most tumor suppressor genes has been confirmed as being due to DNA methylation, and thereby DNA methylation has been a promising biomarker in detecting the disease-associated changes in cells.21 Moreover, DNA methylation is a reversible biochemical modification;22 the transcriptional inactivation of tumor suppressor genes caused by CpG island methylation can be reversed with the DNA methyltransferase inhibitor 5-Aza-CdR, and the reversal of CpG island demethylation can restore the expression of the tumor suppressor gene, allowing the inhibition of cell proliferation and tumor growth.23
In this study, to further explore DNA methylation in CRC, we treated SW480 and LoVo cell lines with 5-Aza-CdR and found that this treatment could reactivate the expression of the epigenetically silenced gene. In addition, our results showed that miR-132 was significantly upregulated after treatment with 5-Aza-CdR, which suggests that DNA hypermethylation is involved in the downregulation of miR-132 expression in CRC. Furthermore, the effect of 5-Aza-CdR on the invasive ability of SW480 and LoVo cells was demonstrated in vitro by transwell assay, revealing that 5-Aza-CdR could inhibit the expression of paxillin by the upregulation of miR-132 and inhibit the invasive ability of CRC cells.
Thus, our study revealed that the suppressive effect of 5-Aza-CdR on the invasive ability of CRC cells may be due to the demethylation and reactivation of miR-132.
Conclusion
In summary, this study suggests that miR-132 binds to the 3′-UTR of paxillin at the posttranscriptional level and is partly downregulated by DNA hypermethylation. Therefore, restoring the expression of miR-132 by using DNA methyltransferase inhibitors has become a new method of cancer gene therapy. The methylation of miR-132 represents a candidate biomarker for further prospective molecular stratification of patients for targeted therapy of CRC.
Acknowledgments
This work was supported by grants from the Jiangsu Province Postdoctoral Science Foundation and the Department of Health of Jiangsu Province of China (number H201214) and project funding from the Affiliated Hospital of Nantong University (number TDFzh201413).
Disclosure
The authors report no conflicts of interest in this work.
References
Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. | ||
Li HZ, Mao WM, Wang XH, Yu CD, Du LB. Incidence and mortality of cancer in Zhejiang province in 2009. Zhonghua Yu Fang Yi Xue Za Zhi. 2013;47:592–596. | ||
Kim J, Huynh R, Abraham I, Kim E, Kumar RR. Number of lymph nodes examined and its impact on colorectal cancer staging. Am Surg. 2006;72:902–905. | ||
Obernosterer G, Tafer H, Martinez J. Target site effects in the RNA interference and microRNA pathways. Biochem Soc Trans. 2008;36:1216–1219. | ||
Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA:target interaction. Chem Biol. 2004;11:1619–1623. | ||
Harfe BD. MicroRNAs in vertebrate development. Curr Opin Genet Dev. 2005;15:410–415. | ||
Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. | ||
Anand S, Majeti BK, Acevedo LM, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16:909–914. | ||
Zheng YB, Luo HP, Shi Q, et al. miR-132 inhibits colorectal cancer invasion and metastasis via directly targeting ZEB2. World J Gastroenterol. 2014;20:6515–6522. | ||
Formosa A, Lena AM, Markert EK, et al. DNA methylation silences miR-132 in prostate cancer. Oncogene. 2013;32:127–134. | ||
Zhang S, Hao J, Xie F, et al. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32:1183–1189. | ||
Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604–610. | ||
Chimonidou M, Strati A, Tzitzira A, et al. DNA methylation of tumor suppressor and metastasis suppressor genes in circulating tumor cells. Clin Chem. 2011;57:1169–1177. | ||
Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42:1273–1281. | ||
Wei X, Tan C, Tang C, et al. Epigenetic repression of miR-132 expression by the hepatitis B virus x protein in hepatitis B virus-related hepatocellular carcinoma. Cell Signal. 2013;25:1037–1043. | ||
Li S, Meng H, Zhou F, et al. MicroRNA-132 is frequently down-regulated in ductal carcinoma in situ (DCIS) of breast and acts as a tumor suppressor by inhibiting cell proliferation. Pathol Res Pract. 2013;209:179–183. | ||
Miller BH, Zeier Z, Xi L, et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc Natl Acad Sci U S A. 2012;109:3125–3130. | ||
Wanet AS, Tacheny A, Arnould T, Renard P. miR-212/132 expression and functions: within and beyond the neuronal compartment. Nucleic Acids Res. 2012;40:4742–4753. | ||
Chen DL, Wang DS, Wu WJ, et al. Overexpression of paxillin induced by miR-137 suppression promotes tumor progression and metastasis in colorectal cancer. Carcinogenesis. 2013;34:803–811. | ||
Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. | ||
Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Mol Biotechnol. 2010;44:71–81. | ||
Ramchandani S, Bhattacharya SK, Cervoni N, Szyf M. DNA methylation is a reversible biological signal. Proc Natl Acad Sci U S A. 1999;96:6107–6112. | ||
Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005;97:1498–1506. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.