Back to Journals » International Journal of Nanomedicine » Volume 16
Downregulation of hsa_circ_0074854 Suppresses the Migration and Invasion in Hepatocellular Carcinoma via Interacting with HuR and via Suppressing Exosomes-Mediated Macrophage M2 Polarization
Authors Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, Li S
Received 29 September 2020
Accepted for publication 11 February 2021
Published 12 April 2021 Volume 2021:16 Pages 2803—2818
DOI https://doi.org/10.2147/IJN.S284560
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ebrahim Mostafavi
Yong Wang,1,* Rongfen Gao,2,* Jinpeng Li,3,* Shaotao Tang,1 Shuai Li,1 Qiangsong Tong,1 Shiwang Li1
1Department of Pediatric Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430022, Hubei, People’s Republic of China; 2Department of Rheumatology and Immunology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430030, Hubei, People’s Republic of China; 3Department of Thyroid and Breast Surgery, Wuhan University Zhongnan Hospital, Wuhan, 430071, Hubei, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yong Wang
Department of Pediatric Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, No. 1277 Jiefang Avenue, Wuhan, Hubei, 430022, People’s Republic of China
Email [email protected]
Background: Circular RNAs (circRNAs) have been identified as key factors in the development of hepatocellular carcinoma (HCC). However, the role and potential molecular mechanism of circRNAs in HCC remain largely unclear. In addition, exosomes are known as important messengers of the cross-talk between tumor cells and immune cells, while the role of extracellular circRNAs in the cell-to-cell communication of tumor cells and immune cells remains not unclear.
Methods: The level of hsa_circ_0074854 in HCC cell lines and HCC cell-derived exosomes was assessed using RT-qPCR assay. In addition, CCK-8 and transwell assays were used to determine the viability, migration and invasion of HCC cells.
Results: Hsa_circ_0074854 expression was upregulated in HCC tissues and HCC cell lines. Additionally, hsa_circ_0074854 knockdown was found to inhibit HCC growth in vitro and in vivo. Mechanistically, hsa_circ_0074854 knockdown inhibited the migration and invasion of HCC cells via interacting with human antigen R (HuR) to reduce its stability. Furthermore, hsa_circ_0074854 can be transferred from HCC cells to macrophages via exosomes. Exosomes with downregulated hsa_circ_0074854 suppressed macrophage M2 polarization, which in turn suppressing migration and invasion of HCC cells both in vitro and in vivo.
Conclusion: Downregulation of hsa_circ_0074854 suppresses the migration and invasion in hepatocellular carcinoma via interacting with HuR and via suppressing exosomes-mediated macrophage M2 polarization. Collectively, these findings may help to understand the diagnosis and treatment of HCC.
Keywords: circular RNAs, hepatocellular carcinoma, exosomes, macrophage polarization, invasion
Introduction
Hepatocellular carcinoma (HCC) is responsible for the third leading cause of cancer deaths worldwide, causing over 600, 000 deaths annually.1,2 Hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, non-alcoholic fatty liver disease, diabetes and alcohol abuse are the important risk factors in the progression of HCC.3–5 Many therapy strategies have been applied to treat HCC, including drug intervention, conservative, surgical and chemoembolization treatment.6 Unfortunately, most HCC patients are often diagnosed in an advanced stage with a worse long-term prognosis, due to delayed diagnosis and treatment.7 However, the five-year survival rate of patients with HCC is >70% if patients are diagnosed at the earliest stage.8 Thus, exploring the promising tumor-related markers may be useful for early diagnosis of HCC.
Circular RNAs (circRNAs) are a group of non-coding RNAs that are characterized by a covalently closed loop structure lacking a cap structure at 5ʹ end and a poly(A) tail at the 3ʹ-end.9,10 CircRNAs were identified to be critical regulators in HCC.11 For instance, circRNA_0000502 inhibited apoptosis and promoted metastasis of HCC cells via targeting microRNA-124.12 Huang et al found that exosomal circRNA-100338 promoted cell metastasis in HCC.13 However, the role and molecular mechanism of circRNAs in HCC need to be further investigated.
Exosomes are small (40–150 nm) extracellular vesicles.14 Exosomes carry a group of specific proteins, lipids, DNA, RNA, miRNA, lncRNA and circRNA and are then transported to adjacent cells or distant cells.15,16 Evidences have been shown that exosomes play a key role in cell-to-cell communication in the tumor microenvironment (TME).17,18 Tumor cell-derived exosomes have been reported to play an important role in promoting tumor invasion and metastasis.19–21 For example, circular RNA IARS can be transported by exosomes from pancreatic cancer cells to endothelial cells, thereby promoting tumor invasion and metastasis.22 However, the relationship between exosomal circRNAs and invasion in HCC remains largely unclear.
Tumor-associated macrophages (TAMs) are the most abundant immune cells in TME to facilitate tumor migration and invasion.23,24 In general, macrophages can be polarized into M1 (classically activated) or M2 (alternatively activated) macrophages depending on environmental cues.25 M1-polarized macrophages are activated by cytokines including IFNγ and LPS, and M2-polarized macrophages are activated by IL-4 and IL-13.26 M1-polarized macrophages play an anti-tumor role, whereas M2-polarized macrophages are thought to promote tumor growth.27 It has been shown that TAMs closely resemble the M2-polarized macrophages.26 Meanwhile, TAMs with an M2 phenotype can promote cancer progression and metastasis.28,29 However, the communication between tumor cells and TAMs remains largely unknown in HCC.
In this study, we found that hsa_circ_0074854 is upregulated in HCC cells, and downregulation of hsa_circ_0074854 inhibited the migration and invasion of HCC cells. Mechanistically, hsa_circ_0074854 knockdown could reduce the stability of human antigen R (HuR) protein and then suppress epithelial–mesenchymal transition (EMT) to inhibit migration and invasion. In addition, oligonucleotide sequences (hsa_circ_0074854-shRNA1) can be transferred by exosomes to macrophages to suppress macrophage M2 polarization. Meanwhile, inhibition of M2-type macrophage of TAMs prevented the migration and invasion of HCC cells in vitro and in vivo. These findings may help to understand the diagnosis and treatment of HCC.
Materials and Methods
Microarray Data Analysis
HCC-related microarray datasets (GSE94508, GSE97332) were downloaded from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/). R language was performed to screen out the differentially expressed circRNAs (DEcircRNAs) between HCC tissues and normal tissues. The circRNAs with P<0.01 and |log2 (FC)|>2 were selected as the significantly DEcircRNAs. The intersection of DEcircRNAs from two datasets was performed using the Venn Diagram package.
Cell Culture
HCC cell lines (HepG2, Huh-7, Hep3B2.1–7, HCCLM3), normal human hepatic cell line L02 and monocytic cell line THP-1 were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cell lines were maintained in DMEM (Thermo Fisher Scientific) containing 10% FBS and kept under 5% CO2 at 37°C. To induce monocyte differentiation into macrophages, THP-1 cells were incubated with phorbol-12 myristate-13 acetate (PMA; 100 ng/mL) for 24 h.
Reverse Transcription Quantitative Real-Time PCR
RNA was extracted from cells using TRIzol reagent (Thermo Fisher Scientific). Complementary DNA (cDNA) was synthesized using an EntiLink™ 1st Strand cDNA Synthesis Kit (Wuhan, China). After that, the qPCR was performed on a StepOne™ Real-Time PCR System (Thermo Fisher Scientific) with the EnTurbo™ SYBR Green PCR SuperMix (Wuhan, China). Primer sequences were listed as in Table 1. β-actin works as an internal control.
 |
Table 1 Primer Sequences |
Lentivirus Infection and Construction of Stable Cell Lines
The oligonucleotides to suppress hsa_circ_0074854 or HuR expression and the oligonucleotides to overexpress HuR expression were designed and obtained by Ribobio (Guangdong, China) respectively. These oligonucleotides were cloned into lentiviral expression vector pLVX-IRES-Puro (Clontech Laboratories). After that, 293 T cells were transfected with vectors described above, and lentivector packaging plasmid mix (pLP/VSVG, pLP1, and pLP2) using Lipo2000 (Thermo Fisher Scientific). Later on, the supernatant containing infectious lentivirus was collected at 48 h, and then filtered through a 0.22 μm filter membrane to obtain viral particles. Next, the viral supernatant was added into the plate to infect HepG2 or Huh-7 cells. Seventy-two hours after transduction, the infected cells were selected by puromycin (2 µg/mL, Amresco, Cleveland, OH, USA).
The oligonucleotide sequences were listed as followed: hsa_circ_0074854-shRNA1: 5ʹ- GGATGCTTCTGGGAATTTATTCAAGAGATAAATTCCCAGAAGCATCCTTTTT-3ʹ; hsa_circ_0074854-shRNA2: 5ʹ-GGCATGTGCAATTGCAGATTTCAAGAGAATCTGCAATTGCACATGCCTTTTT-3ʹ; HuR-shRNA1: 5ʹ-TCGGGAGAACGAATTTGATCGTCAATTCAAGAGATTGACGATCAAATTCGTTCTCCCGATTTTT-3ʹ; HuR-shRNA2: 5ʹ- CGGGATAAAGTAGCAGGACACAGCTTTCAAGAGAAGCTGTGTCCTGCTACTTTATCCCGTTTTT-3ʹ.
Cell Viability Assay
5× 103 HepG2 or Huh-7 cells were plated onto 96-well plates and were incubated for 24 h, 48 h and 72 h. After that, 10 μL of Cell Counting Kit-8 (CCK-8; Dojindo) reagent was added into each well and incubated for another 2 h. Subsequently, the absorbance values at 450 nm were measured using a SpectraMax microtiter plate reader (Molecular Devices).
Cell Apoptosis Assay
Annexin V-FITC Apoptosis Staining/Detection kit (Thermo Fisher Scientific) was utilized to assess cell apoptosis according to the manufacturer’s instructions. Apoptotic cells were analyzed by a flow cytometer (BD Biosciences) using BD FACSDiva software (BD Biosciences).
Western Blot Assay
Enhanced BCA Protein Assay kit (Beyotime) was used to quantify the protein concentration. The protein was separated by SDS-PAGE and then transferred onto polyvinylidene difluoride (PVDF) membranes. After that, the membranes were probed with antibodies against Bcl-2 (1:1000, Abcam Cambridge, MA, USA), active caspase 3 (1:1000, Abcam), PDCD5 (1:1000, Abcam), E-cadherin (1:1000, Abcam), α-SMA (1:1000, Abcam), Vimentin (1:1000, Abcam), HuR (1:1000, Abcam), ZEB-1 (1:1000, Abcam), β-actin (1:1000, Abcam) at 4°C overnight, following by incubation with the corresponding secondary antibodies (1:5000, Abcam) at room temperature for 1 h. Subsequently, the blots were developed by enhanced chemiluminescence (Thermo Fisher Scientific). ImageJ software was used to quantify the protein bands.
RNA Pull-Down Assay
Biotinylated hsa_circ_0074854 was obtained from GenePharma (Shanghai, China). HepG2 cells were transfected with biotinylated hsa_circ_0074854 for 24 h. After that, cell lysates were cultured with streptavidin-coated magnetic beads (Thermo Fisher Scientific). The biotin-coupled RNA complex was pulled down and the level of HuR was detected by RT-qPCR.
Fluorescence in situ Hybridization (FISH)
The FISH assay was performed in HepG2 cells as described previously.30 The fluorescence-conjugated hsa_circ_0074854 or HuR mRNA probes was designed and obtained by RiboBio (Guangzhou, China). Nucleus was counterstained with DAPI. The image was captured by a Leica confocal microscope (Leica Microsystems, Mannheim, Germany).
Exosome Isolation and Characterization
Exosomes were purified from the conditioned medium (CM) of HepG2 cells using ultracentrifugation methods as described previously.31 Briefly, the CM was collected from HepG2 cells, and then centrifuged at 300 × g for 10 min, 2000 × g for 15 min, and 10,000 × g for 30 min to remove cell debris and additional cellular fragments. After that, the supernatants were ultracentrifuged at 120,000 x g for 70 minutes to isolated exosomes. The pelleted exosomes were then resuspended in 100 μL PBS and stored at −80°C. Nanoparticle tracking analysis (NTA) was used to analyze the quantity and size of exosomes. A JSM-7800F transmission electron microscopy (TEM, StarJoy, Japan) was utilized to identify the morphology of exosomes. The HepG2 cell-derived exosomes were analyzed by Western blotting using the following primary antibodies: anti-TSG101 and anti-CD63.
Exosome Labeling and Uptake
Exosomes secreted by the HepG2 cells were fluorescently labeled with PKH26 red membrane dye (Sigma Aldrich). After that, the labeled exosomes were co-cultured with macrophages for 24 h at 37°C. DAPI (blue) was used to label nuclei. Later on, internalization of exosomes by macrophages was observed by a fluorescence microscope (Carl Zeiss, Jena, Germany).
Macrophage Polarization Assay
The macrophages were treated with 20 ng/mL IL-4 and 20 ng/mL IL-13 (for M2 polarization), or HepG2 cell-derived exosomes for 24 h. The macrophages surface makers CD86 (M1 marker) and CD206 (M2 marker) were analyzed by flow cytometry (BD Biosciences).
Transwell Assays
Cell migration and invasion assays were analyzed using 24-well Transwell cell culture chambers (Corning) with or without precoated Matrigel. To explore the effect of hsa_circ_0074854 knockdown on cell migration and invasion, 2 × 104 transfected HCC cells were added into the upper chamber, and the bottom chamber was added with 600 μL medium containing 10% FBS. In addition, to investigate the effect of M2 macrophages induced by HCC cell-secreted exosomes on migration and invasion of HCC cells, 2 × 104 HCC cells were added into the upper chamber. Meanwhile, 1 × 104 macrophages were co-cultured with exosomes, suspended in 600 μL medium with 10% FBS and then added into the bottom chamber. After incubation at 37°C and 5% CO2 for 24 h, the cells on the lower surface of the chamber were stained with crystal violet for 30 min. Subsequently, the migrated and invasive cells were captured using a fluorescence microscope (Leica, Buffalo Grove, IL, USA) in five random microscopic fields, and then the cells were counted.
Animal Studies
BALB/c nude mice (4–5 weeks old) were purchased from the Vital River Laboratories (Beijing, China) and maintained under pathogen-free conditions following the recommended procedures of National Institutes of Health guide for the care and use of laboratory animals. All animal experiments were performed in accordance with a protocol approved by the Ethics Committee of the Union Hospital, Tongji Medical College, Huazhong University of Science and Technology (No. HU20190401X). Animals were divided randomly into seven groups: (I) control, circRNA-shRNA1 and circRNA-shRNA1 + lenti-HuR groups; (II) HepG2, HepG2 + Macrophages (HepG2 + M), HepG2 + exosomes-treated macrophages (HepG2 + MExo) and HepG2 + MExo-circRNA shRNA1 groups. Group I: 5 x 106 HepG2 or HepG2 cells stably expressing hsa_circ_0074854-shRNA1 cells in 100 μL PBS were injected into the left flank of nude mice. Group II: HepG2 cells plus different hsa_circ_0074854 levels exosomes-treated macrophages were subcutaneously injected into left flank of nude mice. The tumor size was measured every week and the volume was calculated by the following formula V = (length x width2)/2. After 3 weeks, the mice were sacrificed, and the tumors were removed. Immunohistochemistry (IHC) was applied to assess CD206 expression in tumor tissues.
Statistical Analysis
Data are presented as the mean ± standard deviation (S.D.). All statistical analyses were performed using GraphPad Prism software (version 7.0, La Jolla, CA, USA). Differences between three or more groups were analyzed by one-way analysis of variance (ANOVA) and Tukey’s tests. Differences between two groups were analyzed by two-tailed Student’s t-test. The differences were considered significant at *P < 0.05. All data were repeated in triplicate.
Results
Identification of DEcircRNAs in the HCC Tissues and Normal Tissues
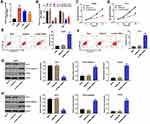
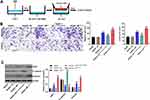
The GSE94508 and GSE97332 datasets were selected for screening the DEcircRNAs between HCC tissues and normal tissues. The upregulated and downregulated circRNAs in HCC tissues compared with normal tissues are shown in Figure 1A and B. A total of 287 and 896 DEcircRNAs were identified in the GSE94508 and GSE97332 datasets, respectively (Figure 1C and D). Using a Venn diagram, 6 overlapping upregulated circRNAs were identified in the GSE94508 and GSE97332 datasets (Figure 1E). The 6 upregulated overlapping DEcircRNAs included hsa_circ_0000517, hsa_circ_0004720, hsa_circ_0003141, hsa_circ_0006913, hsa_circ_00074854, hsa_circ_0088046 (Figure 1F). The highest expression level of hsa_circ_00074854 was observed in HCC tissues.
Hsa_circ_00074854 Knockdown Suppressed Proliferation and Induced Apoptosis of HCC Cells
To investigate the role of hsa_circ_00074854 in HCC cells, we analyzed hsa_circ_00074854 levels in one human hepatic cell line L02, and four HCC cell lines HepG2, Huh-7, Hep3B2.1–7 and HCCLM3, by using RT-qPCR. We found that hsa_circ_00074854 level was notably upregulated in HepG2, Huh-7 and HCCLM3 cells, compared with L02 cells (Figure 2A and Supplementary Figure S1A). Additionally, the level of hsa_circ_00074854 was significantly decreased in HepG2 and Huh-7 cells following transfection with hsa_circ_00074854-shRNA1 (Figure 2B). Moreover, hsa_circ_00074854 knockdown inhibited the proliferation and induced the apoptosis of HepG2 and Huh-7 cells (Figure 2C–F). Meanwhile, downregulation of hsa_circ_00074854 significantly decreased the expression of Bcl-2 and increased the expressions of active caspase 3 and PCDC5 in HepG2 and Huh-7 cells (Figure 2G and H). These results indicated that downregulation of hsa_circ_00074854 could inhibit proliferation and induced apoptosis of HCC cells.
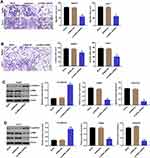
Hsa_circ_00074854 Knockdown Inhibited Migration, Invasion and EMT of HCC Cells
Next, the role of hsa_circ_00074854 in the regulation of migration and invasion in HCC cells was determined using transwell assays. As indicated in Figure 3A and B and Supplementary Figure S1B, downregulation of hsa_circ_00074854 markedly inhibited the migration and invasion abilities of HepG2, Huh-7 and HCCLM3 cells, compared with the shRNA-NC group. In addition, downregulation of hsa_circ_00074854 significantly upregulated the expression of E-cadherin and downregulated the expressions of vimentin and α-SMA in HepG2 and Huh-7 cells (Figure 3C and D). These data suggested that downregulation of hsa_circ_00074854 could inhibit migration, invasion and EMT of HCC cells.
Hsa_circ_00074854 Physically Interacts with HuR in HCC Cells
It has been shown that circRNAs can interact with RNA-binding proteins in cancers.32 HuR, as an RNA-binding protein, has been found to promote migration and invasion of cancer cells.33,34 Thus, we investigated whether hsa_circ_00074854 could regulate HuR in HCC. Then, the interaction between hsa_circ_00074854 and HuR was assessed by RNA pulldown assay. As shown in Figure 4A, the HuR protein was pulled down by hsa_circ_00074854. In addition, FISH results revealed the co-localization of hsa_circ_00074854 and HuR in the cytoplasm of HepG2 cells (Figure 4B). These data indicated a close relationship between hsa_circ_00074854 and HuR.
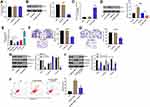
Hsa_circ_00074854 Knockdown Inhibited the Migration, Invasion, and EMT of HepG2 Cells via HuR
We next to investigate the effect of hsa_circ_00074854 on HuR expression. RT-qPCR results showed that hsa_circ_00074854 levels had no change in HuR mRNA levels in HepG2 cells, but Western blot results presented a decreased protein expression of HuR when hsa_circ_00074854 was silenced (Figure 5A and B). In addition, the level of hsa_circ_00074854 was markedly increased in HepG2 cells transfected with lenti-hsa_circ_00074854 (Figure 5C). We then incubated HepG2 cells with the protein synthesis inhibitor cycloheximide (CHX) and assessed the stability of HuR. As shown in Figure 5D, the stability of HuR protein increased when hsa_circ_00074854 was overexpressed and decreased when hsa_circ_00074854 was silenced. To illustrate whether HuR can mediate the migration, invasion and EMT of HepG2 cells, we established HuR knockdown and overexpressed HepG2 cells (Figure 5E). In addition, the results of transwell assays indicated that knockdown of HuR notably suppressed migration and invasion of HepG2 cells (Figure 5F and G). Moreover, downregulation of HuR significantly downregulated the levels of ZEB-1 and vimentin and upregulated the level of E-cadherin in HepG2 cells (Figure 5H). Next, we further investigate whether hsa_circ_00074854 knockdown inhibits the EMT of HepG2 cells via HuR. As indicated in Figure 5I, downregulation of hsa_circ_00074854 notably downregulated the levels of ZEB-1 and vimentin and upregulated the level of E-cadherin in HepG2 cells; however, these levels were markedly reversed by HuR overexpression. Furthermore, downregulation of hsa_circ_00074854 significantly induced the apoptosis of HepG2 cells; however, that effect was reversed by HuR overexpression (Figure 5J). These results suggested that downregulation of hsa_circ_00074854 could inhibit the migration, invasion, and EMT of HepG2 cells via interacting with HuR.
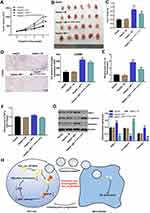
Hsa_circ_00074854 Knockdown Inhibited the Growth of HCC Cells in vivo via HuR
We next investigated whether hsa_circ_00074854 knockdown could affect tumorigenicity in vivo. As revealed in Figure 6A–C and Supplementary Figure S2A–C, downregulation of hsa_circ_00074854 markedly decreased the tumor volume and tumor weight of HepG2 subcutaneous xenografts, compared with control group; however, these effects were markedly reversed by HuR overexpression. These data indicated that hsa_circ_00074854 knockdown could inhibit the growth of HCC cells in vivo.
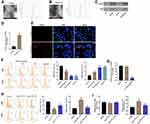
Exosomes with Downregulated hsa_circ_00074854 Suppressed Macrophage M2 Polarization in vitro
Cancer-derived exosomes play an important role in establishing the tumor microenvironment.18 In this study, we isolated exosomes from the conditioned medium (CM) of L02 and HepG2 cells. TEM and NTA were used to identify the determine the quantity and number of exosomes. As shown in Figure 7A and B, exosomes secreted from L02 and HepG2 cells were approximately 100 nm in diameter with typical cup-shaped morphology. In addition, exosomal markers, TSG101 and CD63, were identified using Western blot assay (Figure 7C). These data confirmed that the vesicles isolated from the CM were exosomes. Meanwhile, exosomal hsa_circ_00074854 was found to be enriched in the CM of HepG2 cells, rather than L02 cells, as shown by the RT-qPCR analysis (Figure 7D).
It has been shown that circRNAs can be transferred from tumor cells to recipient cells via exosomes.35 Exosomes present in the tissue microenvironment, and TAMs are the most abundant tumor-infiltrating immune cells in tumor microenvironment.36 Therefore, we hypothesized that cancer-derived exosomal hsa_circ_00074854 participated in the communication between tumor cells and TAMs. To determine whether exosomes could be internalized by recipient cells, PKH26-label exosomes were co-cultured with macrophages (PMA-differentiated human THP-1 monocytes). As revealed in Figure 7E, the uptake of exosomes was observed in macrophages after co-culture for 24 h. In contrast, no PKH26 fluorescence dye was detected in macrophages without the treatment of exosomes (Figure 7E). Evidence has shown that cancer-derived exosomes can induce M2 macrophage polarization.37 Thus, macrophages were incubated with 25 or 50 μg/mL HepG2 cell-derived exosomes (IL-4/IL-13 treatment as a positive control), and the surface markers CD86 (M1 phenotype) and CD206 (M2 phenotype) were analyzed by flow cytometry. As shown in Figure 7F, macrophages exhibited a CD86low/CD206high phenotype, when cells were incubated HepG2 cell-derived exosomes, suggesting that HepG2 cell-derived exosomes could induce the M2 polarization of macrophages. Next, we isolated the exosomes from the CM of HepG2 cells transfected with hsa_circ_00074854-shRNA1 and found that hsa_circ_00074854 levels were significantly decreased in exosomes with downregulated hsa_circ_00074854 (Figure 7G). In addition, macrophages exhibited a CD86high/CD206low phenotype, when cells were incubated with exosomes containing lower hsa_circ_00074854 levels (Figure 7H). Moreover, in comparison with untreated macrophages, macrophages treated with HepG2 cell-derived exosomes expressed markedly more M2 representative cytokine IL-10, but there were no significant changes of M1 representative cytokine TNF-a (Figure 7I). In contrast, macrophages incubated with HepG2 cell-derived exosomes containing lower hsa_circ_00074854 levels showed notably reduced level of IL-10 compared to those incubated with HepG2 cell-derived exosomes (Figure 7I). Overall, exosomes with downregulated hsa_circ_00074854 could suppress macrophage M2 polarization in vitro.
Exosomes with Downregulated hsa_circ_00074854 Inhibited Macrophage M2 Polarization to Suppress Migration, Invasion and EMT in HepG2 Cells in vitro
To explore whether hsa_circ_00074854 knockdown could suppress migration, invasion and EMT of HepG2 cells by suppressing M2 macrophages polarization, macrophages treated with exosomes containing lower hsa_circ_00074854 levels were indirectly co-cultured with HepG2 cells (Figure 8A). The results of transwell migration and invasion assays showed that M2 macrophages induced by HepG2 cell-derived exosomes obviously promoted the migration and invasion of HepG2 cells; however, these effects were markedly reversed by exosomes containing lower hsa_circ_00074854 levels (Figure 8B). In addition, the levels of ZEB-1 and vimentin were increased, and the level of E-cadherin was decreased in HepG2 cells after co-culturing with M2 macrophages induced by HepG2 cell-derived exosomes (Figure 8C). In contrast, macrophages treated with exosomes containing lower hsa_circ_00074854 levels significantly downregulated the levels of ZEB-1 and vimentin and upregulated the level of E-cadherin in HepG2 cells (Figure 8C). Collectively, exosomes with downregulated hsa_circ_00074854 could inhibit macrophage M2 polarization to suppress migration, invasion and EMT in HCC cells.
Exosomes with Downregulated hsa_circ_00074854 Suppressed the Growth of HepG2 Cells via Inhibiting M2 Macrophage Polarization in vivo
To investigate the function of exosomes with downregulated hsa_circ_00074854 in inhibiting macrophage M2 polarization, which suppress the progression of HCC in vivo, HepG2 cells and macrophages treated with exosomes containing lower hsa_circ_00074854 levels were subcutaneously injected into nude mice. As shown in Figure 9A–C, the tumor volume and tumor weight in the HepG2 + MExo-circRNA shRNA1 group was obviously decreased, compared with the HepG2 + MExo group. In addition, macrophages treated with HepG2 cell-derived exosomes upregulated the levels of CD206 and IL-10 in the xenografts; however, these effects were reversed by exosomes containing lower hsa_circ_00074854 levels (Figure 9D–F). Moreover, the level of TNF-a in HepG2 + MExo-circRNA shRNA1 group was not different compared with that in HepG2 + MExo group. Meanwhile, macrophages treated with exosomes containing lower hsa_circ_00074854 levels downregulated the levels of ZEB-1 and vimentin and upregulated the expression of E-cadherin in the xenografts (Figure 9G). These data indicated that hsa_circ_00074854 knockdown inhibited HCC tumorigenesis via suppressing macrophage M2 polarization.
Discussion
CircRNAs have been found to involve in various biological processes, including cell metastasis and invasion.9 In this study, we found that hsa_circ_00074854 is upregulated in HCC tissues. Downregulation of hsa_circ_00074854 inhibited the migration and invasion of HCC cells in vitro and in vivo. Mechanistically, hsa_circ_00074854 knockdown reduced the stability of HuR protein, and then suppressed EMT in HCC cells. Additionally, HCC cell-secreted hsa_circ_00074854 can be delivered to macrophages via exosomes. Meanwhile, exosomes with downregulated hsa_circ_00074854 could inhibit macrophage M2 polarization, thereby suppressing migration, invasion and EMT in HCC cells. These data indicated that hsa_circ_00074854 knockdown could inhibit HCC cell migration and invasion via functioning within cells and affecting the tumor microenvironment.
HuR, an RNA binding protein (RBP), is known to stabilize mRNAs by binding to adenylate/uridylate-rich elements (AREs) and preventing gene degradation.38,39 Evidences have been shown that HuR is highly abundant in many cancers, including HCC.40,41 In addition, elevated HuR expression is associated with poor prognosis of cancer patients.42 Many circRNAs are predicted to interact with RBPs.43 Chen et al found that circAGO2 can bind to HuR to facilitate its activation and then promote cancer progression.44 In this study, RNA pull-down analysis revealed that hsa_circ_00074854 interacted with HuR. Interestingly, when investigating the regulation of hsa_circ_00074854 on HuR expression, we found that hsa_circ_00074854 knockdown decreased the HuR protein level but had no effect on its mRNA level. Meanwhile, we discovered that hsa_circ_00074854 knockdown regulated HuR level in HCC cells via reducing HuR protein stability. Previous study reported that HuR could stabilize ZEB1 mRNA level, and then induce EMT in cigarette smoke extract-treated bronchial epithelial cells.45 Consistently, our results showed that downregulation of HuR decreased the expression of ZEB1 in HCC cells and then inhibited EMT in HCC cells. Meanwhile, hsa_circ_00074854 knockdown could block the ZEB1 signaling, thus suppressing EMT. Collectively, our data indicated that hsa_circ_00074854 knockdown inhibited migration, invasion and EMT in HCC cells via reducing HuR protein stability and inhibition of ZEB1 signaling (Figure 9G).
Our data have found that hsa_circ_00074854 could regulate the proliferation, migration and invasion of HCC cells. In addition, it has been shown that circRNAs can be transported by exosomes from cancer cells to recipient cells within TME.13,46 This process is crucial for tumor progression as well.47 The communication between HCC cells and TME is an important mechanism regulating tumor progression.47 Thus, another important finding from our study was that exosomes with downregulated hsa_circ_00074854 can be delivered to macrophages to inhibit macrophage M2 polarization. Previous study showed that tumor exosomes carry circRNAs and are released into TME.48 TME is a complex system that is composed of a variety of cells and cytokines.49 Importantly, exosomes can create an immune suppression environment for tumor cells to survive via communication between tumor cells and surrounding recipient cells.50 TAMs are the most abundant immune cells in TME, which promote tumor cell migration and invasion mostly by secreting inflammatory cytokines.24,51,52 Meanwhile, M2 macrophage-like TAMs are known to promote tumor proliferation and metastasis.53,54 Evidence has shown that IL-10 secreted by M2 macrophage could promote the migration and invasion of tumor cells.55,56 Consistently, in this study, we found that HCC cell-derived exosomes induced M2 macrophage activation, which promoted migration, invasion and EMT in HCC cells. In contrast, exosomes with downregulated hsa_circ_00074854 suppressed macrophage M2 polarization, which inhibited migration, invasion and EMT in HCC cells both in vitro and in vivo. These data suggested that exosomes with downregulated hsa_circ_00074854 could inhibit macrophage M2 polarization to suppress migration and invasion in HCC cells (Figure 9G). Collectively, exosomes serve as an important messenger that mediated the cross-talk between tumor cells and TAMs.
Conclusion
In summary, we provided the evidence that hsa_circ_0074854 knockdown inhibited the migration and invasion of HCC cells by binding to HuR, thus inhibiting its stability and by suppressing exosomes-mediated macrophage M2 polarization. These findings may help to understand the diagnosis and treatment of HCC.
Data Sharing Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgment
These authors contributed equally to this work and should be considered as co-first authors: Yong Wang, Rongfen Gao, and Jinpeng Li.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Ozer Etik D, Suna N, Boyacioglu AS. Management of hepatocellular carcinoma: prevention, surveillance, diagnosis, and staging. Exp Clin Transplant. 2017;15(Suppl 2):31–35. doi:10.6002/ect.TOND16.L9
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332
3. Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi:10.1038/s41575-019-0186-y
4. Rasool M, Rashid S, Arooj M, et al. New possibilities in hepatocellular carcinoma treatment. Anticancer Res. 2014;34(4):1563–1571.
5. Higashi T, Friedman SL, Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Adv Drug Deliv Rev. 2017;121:27–42.
6. Heimbach JK, Kulik LM, Finn RS, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67(1):358–380. doi:10.1002/hep.29086
7. Ren Z, Ma X, Duan Z, Chen X. Diagnosis, therapy, and prognosis for hepatocellular carcinoma. Anal Cell Pathol. 2020;2020:8157406. doi:10.1155/2020/8157406
8. Tsuchiya N, Sawada Y, Endo I, et al. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2015;21(37):10573–10583. doi:10.3748/wjg.v21.i37.10573
9. Wang L, Long H, Zheng Q, et al. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer. 2019;18(1):119. doi:10.1186/s12943-019-1046-7
10. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
11. Zhang X, Xu Y, Qian Z, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018;9(11):1091. doi:10.1038/s41419-018-1132-6
12. Zhang S, Liu Y, Liu Z, et al. CircRNA_0000502 promotes hepatocellular carcinoma metastasis and inhibits apoptosis through targeting microRNA-124. J BUON. 2019;24(6):2402–2410.
13. Huang XY, Huang ZL, Huang J, et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39(1):20. doi:10.1186/s13046-020-1529-9
14. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–1215. doi:10.1172/JCI81135
15. Dilsiz N. Role of exosomes and exosomal microRNAs in cancer. Future Sci OA. 2020;6(4):Fso465. doi:10.2144/fsoa-2019-0116
16. Li S, Li Y, Chen B, et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46(D1):D106–d112. doi:10.1093/nar/gkx891
17. Atienzar-Aroca S, Flores-Bellver M, Serrano-Heras G, et al. Oxidative stress in retinal pigment epithelium cells increases exosome secretion and promotes angiogenesis in endothelial cells. J Cell Mol Med. 2016;20(8):1457–1466. doi:10.1111/jcmm.12834
18. Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med. 2013;91(4):431–437. doi:10.1007/s00109-013-1020-6
19. Wang H, Lu Z, Zhao X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol. 2019;12(1):133. doi:10.1186/s13045-019-0806-6
20. Ahmadi M, Rezaie J. Tumor cells derived-exosomes as angiogenenic agents: possible therapeutic implications. J Transl Med. 2020;18(1):249. doi:10.1186/s12967-020-02426-5
21. Ghayad SE, Rammal G, Ghamloush F, et al. Exosomes derived from embryonal and alveolar rhabdomyosarcoma carry differential miRNA cargo and promote invasion of recipient fibroblasts. Sci Rep. 2016;6:37088. doi:10.1038/srep37088
22. Li J, Li Z, Jiang P, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37(1):177. doi:10.1186/s13046-018-0822-3
23. Ma YY, He XJ, Wang HJ, et al. Interaction of coagulation factors and tumor-associated macrophages mediates migration and invasion of gastric cancer. Cancer Sci. 2011;102(2):336–342. doi:10.1111/j.1349-7006.2010.01795.x
24. Lan J, Sun L, Xu F, et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019;79(1):146–158. doi:10.1158/0008-5472.CAN-18-0014
25. Wang X, Luo G, Zhang K, et al. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kγ to promote pancreatic cancer metastasis. Cancer Res. 2018;78(16):4586–4598. doi:10.1158/0008-5472.CAN-17-3841
26. Zheng P, Chen L, Yuan X, et al. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res. 2017;36(1):53. doi:10.1186/s13046-017-0528-y
27. Bardi GT, Smith MA, Hood JL. Melanoma exosomes promote mixed M1 and M2 macrophage polarization. Cytokine. 2018;105:63–72. doi:10.1016/j.cyto.2018.02.002
28. Schmall A, Al-Tamari HM, Herold S, et al. Macrophage and cancer cell cross-talk via CCR2 and CX3CR1 is a fundamental mechanism driving lung cancer. Am J Respir Crit Care Med. 2015;191(4):437–447. doi:10.1164/rccm.201406-1137OC
29. Yang L, Dong Y, Li Y, et al. IL-10 derived from M2 macrophage promotes cancer stemness via JAK1/STAT1/NF-κB/Notch1 pathway in non-small cell lung cancer. Int J Cancer. 2019;145(4):1099–1110. doi:10.1002/ijc.32151
30. Huang X, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18(1):71. doi:10.1186/s12943-019-0969-3
31. Greening DW, Xu R, Ji H, Tauro BJ, Simpson RJ. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol. 2015;1295:179–209.
32. Dong W, Dai ZH, Liu FC, et al. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine. 2019;45:155–167. doi:10.1016/j.ebiom.2019.06.030
33. Dormoy-Raclet V, Ménard I, Clair E, et al. The RNA-binding protein HuR promotes cell migration and cell invasion by stabilizing the beta-actin mRNA in a U-rich-element-dependent manner. Mol Cell Biol. 2007;27(15):5365–5380. doi:10.1128/MCB.00113-07
34. Hu YP, Jin YP, Wu XS, et al. LncRNA-HGBC stabilized by HuR promotes gallbladder cancer progression by regulating miR-502-3p/SET/AKT axis. Mol Cancer. 2019;18(1):167. doi:10.1186/s12943-019-1097-9
35. Shi X, Wang B, Feng X, et al. circRNAs and exosomes: a mysterious frontier for human cancer. Mol Ther Nucleic Acids. 2020;19:384–392. doi:10.1016/j.omtn.2019.11.023
36. Yin Z, Ma T, Huang B, et al. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-β signaling pathway. J Exp Clin Cancer Res. 2019;38(1):310. doi:10.1186/s13046-019-1313-x
37. Pritchard A, Tousif S, Wang Y, et al. Lung tumor cell-derived exosomes promote M2 macrophage polarization. Cells. 2020;9(5):1303. doi:10.3390/cells9051303
38. Chatterji P, Rustgi AK. RNA binding proteins in intestinal epithelial biology and colorectal cancer. Trends Mol Med. 2018;24(5):490–506. doi:10.1016/j.molmed.2018.03.008
39. Brennan CM, Gallouzi IE, Steitz JA. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J Cell Biol. 2000;151(1):1–14. doi:10.1083/jcb.151.1.1
40. Kakuguchi W, Nomura T, Kitamura T, et al. Suramin, screened from an approved drug library, inhibits HuR functions and attenuates malignant phenotype of oral cancer cells. Cancer Med. 2018;7(12):6269–6280. doi:10.1002/cam4.1877
41. Zhu H, Berkova Z, Mathur R, et al. HuR suppresses fas expression and correlates with patient outcome in liver cancer. Mol Cancer Res. 2015;13(5):809–818. doi:10.1158/1541-7786.MCR-14-0241
42. Song X, Shi X, Li W, Zhang F, Cai Z. The RNA-binding protein HuR in digestive system tumors. Biomed Res Int. 2020;2020:9656051. doi:10.1155/2020/9656051
43. Hentze MW, Preiss PT. Circular RNAs: splicing’s enigma variations. EMBO j. 2013;32(7):923–925. doi:10.1038/emboj.2013.53
44. Chen Y, Yang F, Fang E, et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26(7):1346–1364. doi:10.1038/s41418-018-0220-6
45. Sun J, Gu X, Wu N, et al. Human antigen R enhances the epithelial-mesenchymal transition via regulation of ZEB-1 in the human airway epithelium. Respir Res. 2018;19(1):109. doi:10.1186/s12931-018-0805-0
46. Bian X, Xiao YT, Wu T, et al. Microvesicles and chemokines in tumor microenvironment: mediators of intercellular communications in tumor progression. Mol Cancer. 2019;18(1):50. doi:10.1186/s12943-019-0973-7
47. Pascut D, Pratama MY, Vo NVT, Masadah R, Tiribelli C. The crosstalk between tumor cells and the microenvironment in hepatocellular carcinoma: the role of exosomal microRNAs and their clinical implications. Cancers (Basel). 2020;12(4):823. doi:10.3390/cancers12040823
48. Shao Y, Lu B. The crosstalk between circular RNAs and the tumor microenvironment in cancer metastasis. Cancer Cell Int. 2020;20:448. doi:10.1186/s12935-020-01532-0
49. Wang J, Li D, Cang H, Guo B. Crosstalk between cancer and immune cells: role of tumor-associated macrophages in the tumor microenvironment. Cancer Med. 2019;8(10):4709–4721. doi:10.1002/cam4.2327
50. Bae S, Brumbaugh J, Bonavida B. Exosomes derived from cancerous and non-cancerous cells regulate the anti-tumor response in the tumor microenvironment. Genes Cancer. 2018;9(3–4):87–100. doi:10.18632/genesandcancer.172
51. Chen Y, Song Y, Du W, et al. Tumor-associated macrophages: an accomplice in solid tumor progression. J Biomed Sci. 2019;26(1):78. doi:10.1186/s12929-019-0568-z
52. Wei C, Yang C, Wang S, et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer. 2019;18(1):64. doi:10.1186/s12943-019-0976-4
53. Yamaguchi T, Fushida S, Yamamoto Y, et al. Tumor-associated macrophages of the M2 phenotype contribute to progression in gastric cancer with peritoneal dissemination. Gastric Cancer. 2016;19(4):1052–1065. doi:10.1007/s10120-015-0579-8
54. Yin M, Li X, Tan S, et al. Tumor-associated macrophages drive spheroid formation during early transcoelomic metastasis of ovarian cancer. J Clin Invest. 2016;126(11):4157–4173. doi:10.1172/JCI87252
55. Qi L, Yu H, Zhang Y, et al. IL-10 secreted by M2 macrophage promoted tumorigenesis through interaction with JAK2 in glioma. Oncotarget. 2016;7(44):71673–71685. doi:10.18632/oncotarget.12317
56. Liu Q, Yang C, Wang S, et al. Wnt5a-induced M2 polarization of tumor-associated macrophages via IL-10 promotes colorectal cancer progression. Cell Commun Signal. 2020;18(1):51. doi:10.1186/s12964-020-00557-2
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.