Back to Journals » Cancer Management and Research » Volume 12
Down-Regulation of Ribosomal Protein RPS21 Inhibits Invasive Behavior of Osteosarcoma Cells Through the Inactivation of MAPK Pathway
Authors Wang T , Wang ZY, Zeng LY, Gao YZ, Yan YX, Zhang Q
Received 22 January 2020
Accepted for publication 27 May 2020
Published 25 June 2020 Volume 2020:12 Pages 4949—4955
DOI https://doi.org/10.2147/CMAR.S246928
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Antonella D'Anneo
Tao Wang,* Zhi-Yong Wang,* Ling-Yuan Zeng, Yao-Zu Gao, Yu-Xin Yan, Quan Zhang
Department of Orthopedics, The Second Hospital of Shanxi Medical University, Taiyuan City, Shanxi Province 030001, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Tao Wang Tel +86-0351-3363078
Email [email protected]
Objective: The goal of our present study was to explore the expression level, biological function, and underlying molecular mechanism of ribosomal protein s21 (RPS21) in human osteosarcoma (OS).
Methods: Firstly, we evaluated the expression of RPS21 in OS tissue samples based on the Gene Expression Omnibus (GEO) datasets and also measured the RPS21 expression of OS cell lines (MG63, and U2OS) by quantitative real-time polymerase chain reaction (qRT-PCR). siRNA interference method was used to reduce the expression of RSP21 in the OS cells. Cell Counting Kit-8 (CCK-8), colony formation, wound-healing, and transwell assays were conducted to measure the proliferation, migration, and invasion of OS cells. The mitogen-activated protein kinase (MAPK) pathway-related proteins levels were examined by Western blot.
Results: Our analyses showed that the expression of RPS21 was significantly increased in OS, compared with normal samples. Upregulation of RPS21 was associated with worse outcomes of OS patients. Knockdown of RPS21 suppressed OS cell proliferation, colony-forming ability, migration, and invasion capacities. Moreover, down-regulation of RPS21 inactivated the MAPK signaling pathway.
Conclusion: RPS21 plays an oncogenic candidate in OS development via regulating the activity of MAPK pathway; therefore, it may serve as a novel therapeutic target for OS treatment.
Keywords: RPS21, MAPK pathway, osteosarcoma, viability, migration
Introduction
Osteosarcoma (OS) is the most frequent bone tumor and occurs in children and adolescents with high incidences.1 Although early diagnosis and treatment have significantly improved the survival rates, the prognosis of OS patients still remains unfavorable, owing to the frequent lung metastasis and the occurrence of multiple drug resistance.2,3 Therefore, it is urgently needed to identify the novel and available signatures to further advance the treatments of OS.
Ribosomal protein s21 (RPS21) is a member of ribosome-associated proteins, which exerts a vital role in ribosome biogenesis, cell growth, and apoptosis.4,5 The increase of ribosome biogenesis is an important characteristic of cell proliferation, and there is an increase of ribosomal activity in tumorigenesis.6,7 Török et al8 have demonstrated that RPS21 is a translation initiation factor and binds to native 40S ribosomal subunits. The disruption of the main components of the translation machinery itself, especially mutation of ribosomal protein genes, can result in cancer.9,10 Significantly, the expression of RPS21 has been implicated in human prostate cancer progression, which might provide an avenue for developing these as the prognostic signature of prostate cancer.11 However, the biological function and potential mechanisms of RPS21 in OS have not been determined so far.
Mitogen-activated protein kinase (MAPK) signaling pathways is an essential intracellular signaling pathway, which has been found to be closely related with the apoptosis of cancer cells,12 including OS.13 Notably, a previous study elucidated that silencing RPS9 suppresses OS cell growth via inactivating MAPK signaling pathway.14 But seriously, whether MPAK signaling pathway participates in the RSP21-mediated the development of OS needs to be investigated.
In our study, RPS21 was remarkably over-expressed in OS. The role of RPS21 in OS cells were further investigated using CCK-8, colony formation, wound-healing and transwell assays. This is the first time to state that RPS21 might function as a prerequisite for the development of OS, and have important prognostic value in OS.
Materials and Methods
Data Mining Relying on GEO Database
Genomics profiling data of Gene Expression Ominibus (GEO) dataset were downloaded to analyze the expressional pattern of RPS21 in OS. Further, GSE16091 array was downloaded to determine the association between RPS21 expression and the outcomes of OS patients.
Cell Lines
The human OS cell lines U2OS and MG63, and the normal osteoblast hFoB1.19 cell line were obtained from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). They were incubated in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA), 100 U/mL penicillin and 100 mg/mL streptomycin (Sigma-Aldrich, St Louis, MO, USA) at 37 °C with 5% CO2.
siRPS21 Transfection
siRPS21 was used to knockdown RSP21 in OS cells and the scrambled siRNA (si-con) were all synthesized by RiboBio Co. Ltd (Guangzhou, China). All resultants were transfected into OS cells using the Lipofectamine 2000 (Invitrogen, Massachusetts, USA) due to the manufacturer’s protocols. Sequences of siRNA targeting human RPS21 were as follows: siRPS21#1: F: 5ʹ- GGUCACAGGCAGGUUUAAU-3ʹ, siRPS21#2: F: 5ʹ- ACUUAUGCUAUCUGCGGGG-3ʹ. After 24 h transfection, the efficiency was examined by means of qRT-PCR and Western blot methods.
Cell Growth Assays
After transfection, Cell Counting Kit-8 (CCK-8) test was conducted to measure the cell proliferation at 0, 24, 48, 72, and 96 h in accordance with the manufacturer’s instructions.
Two weeks later, colonies were fixed in 4% paraformaldehyde, and then dyed with 0.1% crystal violet for 30 min. The colony-forming rate was calculated based on the following formula: (colony number/seeded cell number) x 100%.
Wound Healing Assay
Overall, 5×105 cells were inoculated into six-well plates at 37°C with 5% CO2 for 6 h. A sterile pipette tip was used to create a linear wound in the cellular monolayer, and then cellular debris was removed to yield an acellular area in each well. Images were recorded and the location in the well was noted in terms of the distance between cells.
Transwell Assay
Invasion experiment was implemented by means of a transwell system. The transwell chamber was pre-covered with 250 µg/mL Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). The bottom surface of transwell chamber was filled with RPMI medium with 10% FBS, and serum-free medium was placed on the top surface. Then cells were inoculated into the top surface of transwell chamber at 37°C with 5% CO2 for 24 h-incubation. Cells adhered to the upper surface of the filter were wiped out with a cotton swab, while invaded cells were fixed in 4% paraformaldehyde, and dyed with 1% crystal violet.
For cell migration assay, the transwell chamber without 250 µg/mL Matrigel was prepared. Other steps were similar with the invasion assay. These assays were performed at least three times.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
Total RNA of cells was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA), and then, cDNA was reversely transcribed with a PrimeScript RT Reagent Kit (Takara, Japan). The following primer sequences were applied: RPS21: F: 5ʹ-TCCGCTAGCAATCGCATCAT-3ʹ, R: 5ʹ-GCCCCGCAGATAGCATAAGT-3ʹ; GAPDH: F:5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ, R: 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ. qRT-PCR system was implemented under the following conditions: 95°C x 5 min, 95°C x 30 sec, 60°C x 45 sec, and 72°C x 30 sec. The expression level of RPS21 was normalized to GAPDH. Finally, 2−ΔΔCt method was utilized to calculate the relative expression of RPS21. Three independent experiments were conducted for each group.
Western Blotting
Cell lysate was obtained using TRIZOL reagent based on the standard guidelines of manufacturer. Proteins were separated with 10% SDS-PAGE, and then transferred onto PVDF membranes, and were immunoblotted using primary antibodies including p-ERK, ERK, p-MEK, and MEK. Afterwards, the membranes were incubated using secondary antibody. The protein bands were developed by the ECL reagent (Thermo Fisher Scientific, Rockford, USA).
Statistical Analysis
In this current study, SPSS version 22.0 software (SPSS, Inc., Chicago, IL, USA) was applied to analyze data. All results were presented as the mean ± standard deviation (SD). The differences between two groups was analyzed using the Student’s t-test. One-way ANOVA was utilized to compare the differences among multiple groups. P < 0.05 was regarded as a significant difference.
Results
Analysis of RPS21 Expression Based on the GEO Database
The data sets GSE28424 and GSE36001 were downloaded from GEO database and used to analyze the expression of RPS21 in OS tissues and normal samples. GSE28424 and GSE36001 exhibited that RPS21 expression in OS was higher than that in normal samples (Figure 1A and B, P < 0.05). Moreover, GSE16091 revealed that the highly expressed RPS21 was closely associated with the short survival rate of OS patients (Figure 1C, P < 0.05).
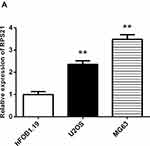
RPS21 Is Remarkably Increased in OS Cells
The endogenous expression of RPS21 in human U2OS, and MG-63 cell lines was investigated using qRT-PCR analysis (Figure 2). As shown in Figure 2, we found that the expression of RPS21 in these OS cell lines was markedly increased, relative to that in the normal osteoblast hFoB1.19 cells. According to that MG63 cells had a higher level of RPS21than U2OS cells, thus, we selected the MG63 cells for the subsequent experiments.
Silencing RPS21 Inhibits Cell Viability of MG63 Cells
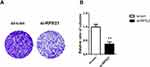
To explore the role of RPS21 in OS, two siRPS21s were used to be transfected into MG63 cells. The transfection efficiency of both siRSP21 #1, and siRSP21#2 was > 80%, not only in mRNA level but in protein level (Figure 3A-C). In addition, higher transfection efficiency of RSP21 expression was observed in the siRSP21#2 group compared with the siRSP21#1 (P > 0.05). Therefore, siRSP21 #2 was used in our follow experiments. After transfection with si-RPS21#2, CCK-8 assay was used to measure the proliferation ability of MG63 cells (Figure 4). As shown in Figure 4, the proliferation of MG63 cells was significantly inhibited at 48, 72, and 96 h (P < 0.05). Furthermore, after knockdown the expression of RPS21, we observed a significant reduction in colony formation (Figure 5A and B, P < 0.05). Collectively, these results showed that RPS21 knockdown may repress the cell growth in OS.
 |
Figure 4 RPS21 knockdown inhibited the proliferation ability of MG63 cells using CCK-8 assay. ** denotes P < 0.01 compared to si-con. |
Knockdown of RPS21 Represses Cell Invasion and Migration in OS Cells
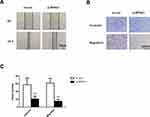
Wound healing assay and transwell analysis were performed to detect the role of RPS21 in the migration and invasion of OS cells. The wound healing assay suggested that knockdown of RPS21 significantly impaired cell migration compared to the control group (Figure 6A, P < 0.05). Transwell assay result demonstrated that down-regulation of RPS21 markedly suppressed tumor invasion and migration potential compared with the control group (Figure 6B and C, P < 0.05). These results demonstrated that the migration and invasion capacities of MG63 cells were significantly decreased by RPS21 suppression in the MG63 cells (Figure 6, P < 0.05).
Reduction of RSP21 Inactivates the MAPK Pathway in MG63 Cells
To attempt to inquire whether RPS21 affects the MAPK pathway, we measured the expression of MAPK signal pathway related proteins using Western blotting, including p-ERK, ERK, p-MEK, and MEK. After si-RPS21knockdown, the expression of p-ERK, and p-MEK were all significantly decreased, but the protein level of ERK, and MEK were not changed (Figure 7A and B, P < 0.05). These observations showed that the promoting-cancer effect of RSP21 in OS was correlated with the activation of MAPK signaling pathway.
Discussion
OS is one of common malignant bone tumors, which seriously affects the healthy of children and adolescents.15 As statistical data described, the five-year survival rate is only 28% for lung metastasis patients.16,17 Thus, this study was attempted to decipher the prospective therapeutic targets for the treatment of OS. After screening the differentially expressed genes based on the GEO datasets, RPS21 was selected as a potential target related with OS progression. RPS21 is a member of ribosome-associated proteins. Over the past decades, emerging evidences have demonstrated that a broader role for dysregulation ribosome biogenesis in the progression of most cancers.18 Besides, a prior study has identified that RPS21 is a candidate diagnostic and prognostic biomarker for prostate cancer.19 Arthurs et al suggested that RPS21 is upregulated in malignant tissues in prostate cancer.20 However, whether RPS21 can affect the development of OS still remains unclear. Therefore, we performed this study to determine the role of RPS21 in OS. On the basis of the GEO dataset, we analyzed the expression of RPS21 in OS as well as its prognostic significance. Result showed that RPS21 was expressed at higher level in OS compared with normal samples. To uncover the internal mechanism, functional in vitro experiments were implemented. These experiments demonstrated that the proliferation and migration capabilities were significantly inhibited in OS cells after RSP21 knockdown. To summarize, RPS21 might function as an oncogenic factor in OS.
As reported, MAPKs regulate a large amount of malignant cellular behaviors, such as proliferation, migration, immune response, and angiogenesis.21 The Ras/Raf/MEK/ERK pathway is active in nearly 30% of human cancers22 and mediates metastasis of many types of cancers.23 Importantly, the clinical benefit of MAPK/ERK-targeted therapy for patients having metastatic or unresectable OS has been reported.24 Significantly, silencing of S100A9 inhibits OS cell proliferation via the inactivation of MAPK signaling pathway.25 Miao et al26 have also suggested that galanin knockdown in MG63 cells hinders cell proliferation as well as invasion capacity through attenuating the activity of MAPK/ERK signaling pathway. Moreover, MEK inhibitors have been reported therapeutic potential in human OS cells.27 Furthermore, a few ribosomal proteins have been found to play vital roles in the progression of OS through mediating MAPK pathway. Highly expressed RPS9 suggests the grave prognosis in OS and its knockdown inhibits OS cells viability probably via inactivation of MAPK signaling pathway.14 RSK2 augments human OS cell growth in vitro via mediating the activity of MAPK pathway.28 Thus, we investigated the effect of RPS21 on the MAPK signaling pathway in OS. Our results revealed that reduction of RPS21 declined the expression levels of p-MEK/p-ERK, which indicated that RPS21 knockdown might inactivate the MAPK pathway in OS cells. Demonstrated herein, the present study illustrated that RPS21 plays an important role in OS through activating the MAPK pathway.
Still, there are some limitations in this study. On the one hand, it is difficult to validate the promoting effect of RPS21 in OS dut to a lack of clinical samples in other independent cohorts, moreover, an in vivo experiment is also needed for more data verification. On the other hand, despite MAPK pathway is changed by RPS21 knockdown, whether it is directly or indirectly involved in RPS21-regulated OS needs to be carefully evaluated in the future.
In summary, for the first time, we found that RPS21 is significantly increased in OS and its upregulation induces poorer prognosis. Moreover, RPS21 inhibition can suppress cell proliferation, migration and invasion of OS cells via the MAPK pathway. These findings provide an experimental basis for deep clarification of the effects of RPS21 on OS and its potential molecular mechanisms. However, we only used OS cell lines to investigate the relationship between RPS21 and OS, but the findings have not been verified in vivo. The only GEO cohort might overstate the RPS21 in OS. Thus, further studies are required to validate the relationship between RPS21 and OS using other data.
Disclosure
The authors declare no conflicts of interest in this study.
References
1. Liu W, Xu G, Liu H, Li T. MicroRNA‐490‐3p regulates cell proliferation and apoptosis by targeting HMGA2 in osteosarcoma. FEBS Lett. 2015;589(20):3148–3153. doi:10.1016/j.febslet.2015.08.034
2. Forman D, Ferlay J, Jemal A, Bray F, Ward E, Center M. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69. doi:10.3322/caac.20107
3. Yu C, Guo Y, Yang H, et al. TRIM66 overexpresssion contributes to osteosarcoma carcinogenesis and indicates poor survival outcome. Oncotarget. 2015;6(27):23708–23719. doi:10.18632/oncotarget.4291
4. Wang Z, Hou J, Lu L, et al. Small ribosomal protein subunit S7 suppresses ovarian tumorigenesis through regulation of the PI3K/AKT and MAPK pathways. PLoS One. 2013;8(11):e79117. doi:10.1371/journal.pone.0079117
5. Maguire BA, Zimmermann RA. The ribosome in focus. Cell. 2001;104(6):813–816. doi:10.1016/S0092-8674(01)00278-1
6. Xiang Z, Wen-Juan L, Jun-Ming L, Peng L, Hua L. Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol. 2015;7(2):92–104. doi:10.1093/jmcb/mjv014
7. Lai MD, Xu J. Ribosomal proteins and colorectal cancer. Curr Genomics. 2007;8(1).
8. Török I, Herrmannhorle D, Kiss I, et al. Down-regulation of RpS21, a putative translation initiation factor interacting with P40, produces viable minute imagos and larval lethality with overgrown hematopoietic organs and imaginal discs. Mol Cell Biol. 1999;19(3):2308. doi:10.1128/MCB.19.3.2308
9. Lai K, Amsterdam A, Farrington S, Bronson RT, Hopkins N, Lees JA. Many ribosomal protein mutations are associated with growth impairment and tumor predisposition in zebrafish. Develop Dynamics. 2009;238(1):76–85. doi:10.1002/dvdy.21815
10. Amsterdam A, Sadler KC, Lai K, et al. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2(5):E139. doi:10.1371/journal.pbio.0020139
11. Arthurs C, Murtaza BN, Thomson C, et al. Expression of ribosomal proteins in normal and cancerous human prostate tissue. PLoS One. 2017;12(10):e0186047. doi:10.1371/journal.pone.0186047
12. Kuo WH, Chen JH, Lin HH, Chen BC, Hsu JD, Wang CJ. Induction of apoptosis in the lung tissue from rats exposed to cigarette smoke involves p38/JNK MAPK pathway. Chem Biol Interact. 2005;155(1–2):31–42. doi:10.1016/j.cbi.2005.04.008
13. Chen HJ, Lin CM, Lee CY, et al. Kaempferol suppresses cell metastasis via inhibition of the ERK-p38-JNK and AP-1 signaling pathways in U-2 OS human osteosarcoma cells. Oncol Rep. 2013;30(2):925–932. doi:10.3892/or.2013.2490
14. Cheng DD, Zhu B, Li SJ, Yuan T, Yang QC, Fan CY. Down-regulation of RPS9 inhibits osteosarcoma cell growth through inactivation of MAPK signaling pathway. J Cancer. 2017;8(14):2720–2728. doi:10.7150/jca.19130
15. Lucero CMJ, Vega OA, Osorio MM, et al. The cancer-related transcription factor runx2 modulates cell proliferation in human osteosarcoma cell lines. J Cell Physiol. 2013;228(4):714–723. doi:10.1002/jcp.24218
16. Yan GN, Lv YF, Guo QN. Advances in osteosarcoma stem cell research and opportunities for novel therapeutic targets. Cancer Lett. 2016;370(2):268. doi:10.1016/j.canlet.2015.11.003
17. Mio H, Mitsuhiko O, Yusuke K, et al. PAI‐1, a target gene of miR‐143, regulates invasion and metastasis by upregulatingMMP‐13 expression of human osteosarcoma. Cancer Med. 2016;5(5):892–902. doi:10.1002/cam4.651
18. Pelletier J, Thomas G, Volarević S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer. 2018;18(1):51–63. doi:10.1038/nrc.2017.104
19. Liang Z, Mou Q, Pan Z, et al. Identification of candidate diagnostic and prognostic biomarkers for human prostate cancer: RPL22L1 and RPS21. 2019;36(6):56. DOI:10.1007/s12032-019-1283-z
20. Arthurs C, Murtaza BN, Thomson C, et al. Expression of ribosomal proteins in normal and cancerous human prostate tissue. PLoS One. 2017;12(10):e0186047. DOI:10.1371/journal.pone.0186047
21. Hu JY, Chu ZG, Han J, et al. The p38/MAPK pathway regulates microtubule polymerization through phosphorylation of MAP4 and Op18 in hypoxic cells. Cell Mol Life Sci. 2010;67(2):321. doi:10.1007/s00018-009-0187-z
22. Boldt S, Kolch W. Targeting MAPK signalling: prometheus’ fire or Pandora’s box? Curr Pharm Des. 2004;10(16):1885. doi:10.2174/1381612043384420
23. Young A, Lyons J, Miller AL, Phan VT, Alarcón IR, Mccormick F. Ras signaling and therapies. Adv Cancer Res. 2009;102:1.
24. Chandhanayingyong C, Kim Y, Staples JR, Hahn C, Lee FY. MAPK/ERK signaling in osteosarcomas, ewing sarcomas and chondrosarcomas: therapeutic implications and future directions. Sarcoma. 2012;2012(25):404810. doi:10.1155/2012/404810
25. Cheng S, Zhang X, Huang N, Qiu Q, Jin Y, Jiang D. Down-regulation of S100A9 inhibits osteosarcoma cell growth through inactivating MAPK and NF-κB signaling pathways. BMC Cancer. 2016;16(1):253. doi:10.1186/s12885-016-2294-1
26. Miao JH, Wang SQ, Zhang MH, et al. Knockdown of galectin-1 suppresses the growth and invasion of osteosarcoma cells through inhibition of the MAPK/ERK pathway. Oncol Rep. 2014;32(4):1497–1504. doi:10.3892/or.2014.3358
27. Baranski Z, Booij TH, Kuijjer ML, et al. MEK inhibition induces apoptosis in osteosarcoma cells with constitutive ERK1/2 phosphorylation. Genes Cancer. 2015;6(11–12):503–512. doi:10.18632/genesandcancer.91
28. Qiu Q, Jiang J, Lin L, et al. Downregulation of RSK2 influences the biological activities of human osteosarcoma cells through inactivating AKT/mTOR signaling pathways. Int J Oncol. 2016;48(6):2508–2520. doi:10.3892/ijo.2016.3481
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.