Back to Archived Journals » Advances in Genomics and Genetics » Volume 5
Double-strand break repair mechanisms in Escherichia coli: recent insights
Authors Đermić D
Received 16 October 2014
Accepted for publication 3 December 2014
Published 9 January 2015 Volume 2015:5 Pages 35—42
DOI https://doi.org/10.2147/AGG.S51699
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr John Martignetti
Damir Ðermić
Ruđer Bošković Institute, Division of Molecular Biology, Zagreb, Croatia
Abstract: In order to survive, all organisms must repair the continuous appearance of double-strand breaks (DSBs) in their DNA. Escherichia coli does this by RecA-dependent homologous recombination (HR), during which the RecA protein is assembled on a 3´-terminated overhang that is created by a process called DNA end resection. The RecA nucleoprotein filament searches for and invades an intact homologous DNA sequence, creating a central HR intermediate. This review describes recent insights into HR and DSB repair in E. coli, especially the processes that precede the formation of a RecA nucleoprotein filament, with an emphasis on the regulation of 3´-tail metabolism. Since HR is a highly conserved process, the parallels to DSB repair in eukaryotic systems are discussed, bearing in mind that the lessons learned from studies in simpler bacterial models may be useful for studying DSB repair and the maintenance of genome stability in eukaryotes.
Keywords: RecA nucleoprotein filament, homologous recombination, exonucleases, genome stability, 3´-overhang metabolism
Introduction
Among the factors that threaten genomic stability and integrity, DNA double-strand breaks (DSBs) are particularly lethal. Induced by exogenic (ultraviolet [UV], ionizing radiation [IR], etc) or endogenic (alterations in DNA metabolic reactions: replication; repair; etc) sources, DSBs must be repaired in all living organisms in order for chromosome replication and transcription to proceed. DSBs are eliminated predominantly by nonhomologous end joining (NHEJ) and homologous recombination (HR). During NHEJ, two ends of a break are aligned and ligated, thus restoring DNA continuity without requiring an intact copy of the affected locus though, in effect, causing mutations. Yet, the most universally distributed DSB repair pathway is HR, mediated by evolutionarily conserved recombinase proteins (RecA, RadA, and Rad51 [Dmc1] from bacteria, archaea, and eukaryotes, respectively), which repair a broken chromosome using an intact copy as a template in an error-free process.
In Escherichia coli, HR is the exclusive DSB repair pathway during which the RecA recombinase binds single-strand (ss)DNA derived from a break, thus creating a RecA nucleoprotein filament (the main recombination intermediate). The nucleoprotein filament subsequently performs a homology search and exchanges the broken DNA sequence with its intact homologue.1 Since recombinases have lower affinity for ssDNA than their cognate ssDNA-binding proteins, SSB/RPA, recombinase polymerization on ssDNA is mediated by a recombination mediator class of proteins in a regulated manner.2 These are the RecBCD and RecFOR proteins in bacteria, and BRCA2, PALB2, and Rad52 in eukaryotes.2 The critical step in recombinase–nucleofilament assembly is the creation of a ssDNA fragment in broken DNA, onto which the recombinase can bind. DSBs are thus processed by a combination of helicase/nuclease activities that lead to 3′-end overhangs, a substrate for recombinase binding. This process involves nucleolytic degradation of a complementary 5′-strand and is referred to as end resection.3 There are several pathways of DNA end resection in E. coli, and these have been traditionally used to define the HR pathways.1
RecBCD pathway and its variants
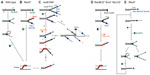
In wild-type E. coli, a multifunctional heterotrimeric RecBCD enzyme repairs a great majority of DSBs.1 It binds blunt or nearly blunt double-strand (ds)DNA ends with high affinity and, with a combination of fast and processive helicase and nuclease activities, unwinds and degrades both strands of a DNA duplex (Figure 1A).4 RecB and RecD subunits are helicases, with RecB also having a unique nuclease and RecA-loading domain.5–8 The destructive mode of RecBCD’s action (which is useful for destroying foreign DNA) is converted to a productive mode by the enzyme’s interaction with a regulatory octanucleotide DNA sequence Chi, whereupon the enzyme continues unwinding DNA and degrading a 5′-terminated strand, while leaving a 3′-post-Chi strand intact.9 Moreover, Chi-modified RecBCD starts facilitating RecA loading onto such a created 3′-overhang.10 In this way, the coupled helicase/nuclease/RecA-loading activities of RecBCD are sufficient for creating a RecA nucleoprotein filament in broken DNA.
In recD null mutants, DSBs are efficiently repaired by the RecBC enzyme even though it has no nuclease activity and does not interact with a Chi sequence.1 RecBC unwinds the DNA duplex and loads RecA constitutively onto the 3′-tail (hence, behaving similarly to the Chi-modified RecBCD, except that it lacks 5'–3′ exonuclease activity) (Figure 1B).11,12 The RecBC’s missing 5′–3′ exonuclease activity is effectively compensated for by the activity of RecJ and exonuclease (Exo)VII, cellular exonucleases that degrade ssDNA (ssExos) from a 5′-end.13 RecJ is the dominant 5′–3′ ssExo with ExoVII (which also possesses 3′–5′ exonuclease activity) providing backup activity.13 The importance of trimming the 5′-strand of a dsDNA end is evident in a recD mutant deficient in RecJ and ExoVII, which is barely viable and is recombination deficient, as well as extremely sensitive to UV and gamma irradiation.13,14
Another RecBCD-derived pathway is operative in recB1080 (and recB1067) mutants, which produce the RecB1080CD (RecB1067CD) enzyme. A single amino acid substitution in the active center of the RecB nuclease domain renders these enzymes nuclease and RecA-loading deficient, while leaving their DNA binding and helicase activities unaffected.7,15 In these cells, RecB1080CD provides rapid and processive helicase activity, while RecJ exonuclease and RecFOR proteins perform 5′–3′ exonuclease and RecA-loading activities, respectively (Figure 1C).16,17 The resulting HR and DNA repair processes in the recB1080 mutant are less efficient, which results in its lower viability and increased sensitivity to DNA-damaging agents when compared to wild-type bacteria.16
The nuclease domain of the RecB subunit is connected to the helicase domain by a long linker,18 which contributes to the flexibility in its substrate specificity. The RecBCD nuclease domain degrades both of the unwound strands; however, it switches to degradation of just the 5′-terminated strand upon the enzyme’s interaction with a Chi site in the DNA. Also, the RecBC enzyme assumes a type of conformation that renders it nuclease deficient, even though it possesses an intact nuclease domain.
RecF pathway
In contrast to RecBCD’s activity on DSBs, RecFOR proteins in wild-type E. coli mediate the polymerization of RecA on internal ssDNA regions or gaps, which originate mainly from replication defects.19 E. coli mutants lacking the RecBCD enzyme are poorly viable and are highly sensitive to genotoxic agents that induce DSBs, unless ssExos of proven (ExoI) and apparent (SbcCD) 3′–5′ polarity are inactive.1 In this genetic background, a RecQ helicase and, to a lesser extent, a UvrD helicase unwind break ends (a role for a HelD helicase has been ruled out recently),20 thus enabling RecJ-catalyzed degradation of the unwound 5′-overhang, while its 3′-complement is bound by RecA with the help of RecFOR proteins (Figure 1D).1 Hence, inactivation of ExoI and SbcCD ssExos allows for the combined activity of several helicases/nucleases/recombination mediator proteins to effectively replace the RecBCD function in DSB repair. This demonstrates the robustness of the E. coli DSB repair capability, which is not surprising considering the importance of DSB removal for cell survival.
A RecA nucleoprotein filament formed by any of the aforementioned pathways searches for an intact homologous sequence and invades it, thus creating a displacement loop (also called a D-loop) structure.1 Ensuing strand exchange reactions and replication initiated by PriA turn this three-strand recombination intermediate into a replication fork,21 when a single dsDNA end (eg, a collapsed replication fork) is repaired. Alternatively, when two dsDNA ends of a DSB are processed, they give rise to a pair of four-strand structures designated as Holliday junctions. Finally, the HR process is completed by resolving the Holliday junctions through the actions of the RuvABC resolvase,22 thus producing mature recombination products.
Real-time monitoring of DSB repair in E. coli
By using an I-SceI endonuclease system for chromosomal DSB induction in combination with fluorescently marked proteins, a recent study23 has characterized the progression of DSB repair in live E. coli cells. This revealing study has shown that a DSB in a site-specific chromosome locus is repaired using distant sister homology as a template. In wild-type cells, RecBCD processing of both ends of a DSB precedes RecA polymerization, which nucleates into large structures that the authors term bundles.23 Once formed, two RecA bundles quickly relocate toward uncut homologous loci, with which they pair.23 Throughout all of these reactions, the two ends of a DSB are kept in close proximity, suggesting that both of them participate in DSB repair, as seen in HR-dependent DSB repair in eukaryotes.24 The study revealed that RecN, a relatively poorly characterized DSB repair protein, is recruited to a DSB early, which is in accord with an earlier study showing that a RecA nucleofilament mobilizes RecN.25 In addition, as expected, PriA and RecG were shown to be recruited to a DSB later, thus confirming that their functions in replication restart and processing of Holliday junctions, respectively, are required for DSB repair.23 Lastly, the kinetics of DSB repair in E. coli was determined:23 RecBCD degradation of the ends of a DSB lasts for ~60 minutes, during which time RecA bundles form starting at ~5 minutes postcut, and reaching a maximum at ~15 minutes postcut. Bundle relocation lasts for ~47 minutes, followed by sister loci pairing, which takes ~5 minutes; then, in another ~17 minutes, the bundles are disassembled. Sister pairing persists for ~50 minutes before their segregation. Overall, it takes ~150 minutes to repair a DSB in wild-type E. coli.23 However, as the cells in this study were grown at 30°C and microscopy was performed at 24°C, it is likely that DSB repair takes less than 150 minutes at the physiological temperature of 37°C.
A slight change in RecA protein strongly increases E. coli resistance to high doses of ionizing radiation
When E. coli was repeatedly subjected to 3,000 Gy of IR in directed evolution experiments, after 20 such cycles, the survivors became 3–4 orders of magnitude more radioresistant than the initial population.26 recA is one of the three genes involved in DNA metabolism whose mutation leads to a majority of the extreme resistance.27 Proteins produced by such mutated recA alleles, RecAD276N and RecAD276A, increase IR resistance of the starter strain by approximately tenfold.27 Biochemical characterization of the two altered proteins revealed that they are less sensitive to inhibition by adenosine diphosphate. They also nucleate a RecA nucleoprotein filament more rapidly, but extend it more slowly than wild-type RecA.27 As a result, a higher number of shorter RecA nucleoprotein filaments can be formed in a cell. This is certainly beneficial for the repair of a large number of DSBs caused by 3,000 Gy of IR. This example shows that E. coli has the capacity to repair many more DSBs than it normally does, with just slight alterations (a single amino acid change in this case) in the existing repair pathways. However, as E. coli hardly ever encounters such high levels of IR, wild-type RecA is optimized for repairing a low number of simultaneous DSBs in a cell, for which longer RecA nucleofilaments might be favorable because they enable more rapid homologous DNA pairing.28 In this respect, one would expect that the viability and DNA repair efficiency of the recA276 mutants exposed to low doses of genotoxic agents is worse than those of their parental wild-type cells, and also that the recA276 mutants would be outgrown in competition assays with wild-type bacteria when IR is not applied.
3′-overhang metabolism matters in E. coli DSB repair
While the significance of nucleolytic degradation of 5′-terminated strands emanating from a DSB is well appreciated, DNA end resection is defined by it; recent studies, which will be discussed, have pointed out the importance of complementary 3′-end strand metabolism in HR. Several aspects of 3′-end metabolism depict DNA end resection as a rather complicated process, which cannot be confined to just 5′-strand degradation.
The balance of helicase and exonuclease activities in HR
In RecBCD-deficient cells, as described in the RecF pathway section, the inactivation of 3′–5′ ssExos, ExoI and SbcCD, enables DSB repair, with RecQ and RecJ providing 3′–5′ helicase and 5′–3′ exonuclease activity, respectively. RecQ is a weak helicase (unwinds ~2 base pairs per second)29 and it certainly cannot compete with the strong, processive 3′–5′ ssExos ExoI (trims ~275 nucleotides per second)30 and SbcCD; hence, no HR occurs in RecBCD− ExoI+ SbcCD+ cells. However, Buljubašić et al20 have recently shown that in RecBCD− ExoI− SbcCD− mutant, the additional inactivation of ExoVII, a processive 3′–5′ ssExo,31 relieves the requirement for the RecQ helicase in HR. On the other hand, if a distributive 3′–5′ ssExo ExoX32 is inactivated instead of ExoVII, the RecQ requirement is not alleviated.20 Hence, in recBCD mutants, a requirement for helicase activity in DSB repair depends on the amount of 3′–5′ exonuclease activity – ie, the weaker the 3′–5′ exonuclease activity is, the less the requirement for the helicase activity. An in vitro RecF pathway reconstruction study33 has revealed similar effects; namely, in experiments lacking 3′–5′ ssExos, RecQ is loosely required and RecJ alone can produce long enough ssDNA from a dsDNA end to enable a RecA-catalyzed strand invasion, albeit at lower efficiency than in a RecQ-assisted reaction. This means that in the absence of 3′–5′ exonuclease activity, helicase activity becomes dispensable for the RecF pathway of HR due to RecJ’s ability to degrade dsDNA.33 However, since HR and DNA repair are very efficient in RecBCD− ExoI− SbcCD− ExoVII− RecQ− UvrD− mutants,20 wherein a 3′-tail is disposed to the action of 3′–5′ ssExos (eg, ExoX, PNPase,34 RNase T,35 etc), it is quite likely that some other helicase(s) process(es) dsDNA ends in these cells.
The aforementioned is a situation where the combination of weak helicase and strong 3′–5′ exonuclease activity prevents HR. Another study36 has recently described the opposite situation, in which a helicase is so strong that it outperforms the 3′–5′ ssExos; this situation is also deleterious for a cell. Namely, in the recB1080 mutant, a highly processive (estimated at ≥50 kbp/binding event) and fast (1–2 kbp/s) RecB1080CD helicase produces tens of kb long overhangs.36 Cell viability, DNA repair, and HR of the mutant are impaired when compared to the wild-type strain, with further deterioration observed upon inactivation of 3′–5′ ssExos ExoI,36 ExoVII, SbcCD, and ExoX (Đermić, unpublished data). A deleterious effect of increased processivity of DNA end-processing, resulting in longer 3′-overhangs, was noted in eukaryotic cells as well, which showed increased genomic instability and higher sensitivity to DNA damage.37–39
Together, these situations show that the coordination of helicase and 3′–5′ exonuclease activity is essential for efficient DSB processing in E. coli. An imbalance in that coordination may result in either an inability to create a 3′-overhang (when the 3′–5′ exonuclease activity is dominant), or in the production of a very long 3′-tail (when helicase activity dominates), which is reactive and tends to engage in aberrant, nonproductive reactions (which will be discussed). This argument provides the rationale for the coupling of robust nuclease activity to strong helicase activity in the RecBCD enzyme. From that perspective, RecBCD nuclease activity’s raison d’être is primarily DSB repair rather than degradation of foreign DNA. Many phages have developed protection mechanisms against RecBCD, while bacteria have developed RecBCD-independent adaptive immunity against invasive exogenic genetic elements.40 During DSB repair, RecBCD nuclease activity prevents long 3′-terminated overhangs from appearing in a cell, while continuously removing 5′-strands. Indeed, in the RecBCD machine, the fast and processive helicase activity is coupled to powerful exonuclease activity on both unwound strands until interaction with a Chi site marks the DNA duplex as a host molecule that should be repaired instead of destroyed. At that point the Chi-modified enzyme slows down and stops degradation of the 3′-strand while continuing degradation of the 5′-strand.1,4 Although the Chi-modified RecBCD is slowed down, the recombination reactions it catalyzes are insensitive to ExoI, the strongest 3′–5′ ssExo,36 thus suggesting that the enzyme has optimally coordinated helicase and nuclease activities. In addition, the polymerized RecA protein may protect a post-Chi 3′-overhang from the activity of ExoI.11
Remarkably, there is an analogous example of balancing helicase and nuclease activities in the eukaryotic end-resection protein, Dna2, which possesses both 5′–3′ helicase and 5′–3′ nuclease activity.41 During end resection, the nuclease activity of this RecB related protein is coupled to the helicase activity of the RecQ-like helicases, Sgs1 and BLM, while its own helicase activity is not required.42,43 Saccharomyces cerevisiae Dna2 deficient in nuclease activity is a vigorous helicase, suggesting that its nuclease activity suppresses its helicase activity.44 A structural switch in Dna2 that regulates the balance between its helicase and nuclease activities remains to be determined.
Coordination of helicase and RecA-loading activity in RecBCD determines the efficiency and fidelity of DSB repair
In addition to being a kind of “runaway” helicase devoid of nuclease activity, RecB1080CD is also deficient in the facilitation of RecA loading.15 That makes it more akin to the DNA end resection machines in eukaryotes, which also have uncoupled helicase/nuclease and recombinase-loading activities (unlike RecBCD).36 The recB1080 mutant is thus a convenient E. coli model for studying eukaryotic DNA end resection mechanisms.36 In this respect, it is interesting that inactivation of the RecQ helicase impairs cell viability, DNA repair, and HR of the recB1080 mutant, thus resembling sickly phenotypes caused in eukaryotes by deficiencies in RecQ’s eukaryotic analogs.36 Also, the deleterious effect of ExoI inactivation in the recB1080 mutant, as described earlier, is exacerbated by inactivation of the RecQ helicase.36 A recB1080 mutant lacking ExoI and RecQ is poorly viable, HR and DNA repair deficient and, notably, displays highly increased illegitimate recombination.36 The deleterious effects of ExoI and RecQ inactivation are not observed in wild-type and recD genetic backgrounds,36 which display the helicase/nuclease/RecA-loading and helicase/RecA-loading activities of RecBC(D), respectively. Therefore, it may be inferred that the combined helicase and RecA-loading activity of the RecBC(D) enzyme funnels 3′-overhangs into productive HR reactions and away from aberrant, nonproductive reactions, thus preventing illegitimate recombination.36
In addition to its role in the initiation of the RecF pathway of HR, RecQ also binds to recombination intermediates, such as Holliday junctions and joint molecules in Kappa and D-loop formation,45 which it disrupts, especially those arising from aberrant, nonhomologous joint exchanges.46,47 RecQ and its widespread orthologs are structurally and functionally highly conserved, and members of this protein family participate in both HR initiation and disruption/regulation, thus acting as key genome caretakers from bacteria to humans.47,48
In the recB1080 ExoI− RecQ− mutant, long 3′-overhangs produced by the fast and processive RecB1080CD helicase have increased longevity due to ExoI inactivation, and they are also more reactive because of RecQ deficiency (Figure 1C), which is very deleterious for the cell.36 These effects show that the metabolism of the 3′-terminated strands is seemingly equally as important as degradation of the 5′-terminated strands for efficient and precise DSB repair in E. coli and, hence, for its genome stability.
Interestingly, RecBCD’s Bacillus subtilis counterpart, AddAB, does not facilitate RecA polymerization on a 3′-tail, which is catalyzed by RecOR mediators instead.49 The AddAB class of helicase/nuclease proteins is present in considerably more bacterial species than the RecBCD class.50 This means that intimate coupling of helicase and recombinase-polymerizing activities occurs in the DSB end resection apparatus of only a minority of bacteria, while eukaryotic organisms, archaea, and the majority of bacteria have looser coordination between the two activities and, consequently, they have evolved different mechanisms for assuring precise and efficient DSB repair. Therefore, it is likely that due to uncoupling of the helicase and RecA-loading activity of AddAB (analogous to RecB1080CD), a role of 3′–5′ ssExos and RecQ helicase (and RecOR proteins) is more prominent in preserving genome stability in bacterial species that harbor AddAB than in those harboring the RecBCD enzyme.
3′–5′ ssExos control DSB processing in a recA mutant
Although a recA null mutant is HR deficient, the RecBCD enzyme nevertheless helps maintain its viability.4 This beneficial role is due to RecBCD’s exonuclease activity, which is in fact unregulated, “reckless”, and may lead to the degradation of a complete chromosome.51 It has recently been shown that RecBCD-catalyzed DNA degradation in the recA mutant is dependent on 3′–5′ ssExos ExoI, SbcCD, and ExoVII.52,53 Genetic evidence suggests that these ssExos are required for loading and/or reloading of the main DSB processing machine RecBCD to dsDNA ends in recA cells (Figure 1E).52,53 And since RecBCD loading to DNA is not dependent on ExoI, SbcCD, and ExoVII in wild-type cells,52,53 one can conclude that dsDNA ends in recA cells, unlike those in wild-type bacteria, contain long 3′-overhangs that prevent RecBCD loading (Figure 1E). Two sources for such ends occurring in recA cells are proposed: a regressed replication fork would give rise to a dsDNA end with a long 3′-tail in the absence of the RecA protein; and/or a post-Chi 3′ tail created by RecBCD in recA bacteria is not covered by the RecA protein and is therefore not immobilized in a HR intermediate, nor is it protected from degradation by ExoI, and possibly other 3′–5′ ssExos.52,53
The aforementioned role of the SbcCD exonuclease in enabling loading of the main DSB processing machine to DSBs is especially interesting, as it mimics the role of SbcC and SbcD eukaryotic orthologs, Rad50 and Mre11, respectively, in bringing about recruitment of the main DSB processing proteins that perform long-range DSB end resection.3 This, in turn, directs DSB repair toward HR and, to a lesser extent, microhomology-mediated end joining repair pathways, and away from NHEJ.54 However, the difference between bacterial SbcCD and its eukaryotic equivalents is that the former participates in trimming of 3′-tails (as suggested by ample genetic evidence),13,20,51,55 whereas the latter performs initial short-range DSB end resection by degrading 5′-strands, even though it also possesses exonuclease activity of the 3′–5′ polarity.56
Acknowledgments
The author is supported by Croatian Science Foundation, project 2978. I am grateful to Nikola Paić and Mary Sopta for their help with manuscript preparation.
Disclosure
The author reports no conflicts of interest in this work.
References
Persky NS, Lovett ST. Mechanisms of recombination: lessons from E. coli. Crit Rev Biochem Mol Biol. 2008;43(6):347–370. | |
Zelensky A, Kanaar R, Wyman C. Mediators of homologous DNA pairing. Cold Spring Harb Perspect Biol. 2014;6(12). pii: a016451. | |
Symington LS. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb Perspect Biol. 2014;6(8). pii: a016436. | |
Yeeles JT, Dillingham MS. The processing of double-stranded DNA breaks for recombinational repair by helicase-nuclease complexes. DNA Repair (Amst). 2010;9(3):276–285. | |
Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423(6942):889–893. | |
Dillingham MS, Spies M, Kowalczykowski SC. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423(6942):893–897. | |
Yu M, Souaya J, Julin DA. Identification of the nuclease active site in the multifunctional RecBCD enzyme by creation of a chimeric enzyme. J Mol Biol. 1998;283(4):797–808. | |
Spies M, Kowalczykowski SC. The RecA binding locus of RecBCD is a general domain for recruitment of DNA strand exchange proteins. Mol Cell. 2006;21(4):573–580. | |
Anderson DG, Kowalczykowski SC. The recombination hot spot Chi is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 1997;11(5):571–581. | |
Anderson DG, Kowalczykowski SC. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a Chi-regulated manner. Cell. 1997;90(1):77–86. | |
Churchill JJ, Anderson DG, Kowalczykowski SC. The RecBC enzyme loads RecA protein onto ssDNA asymmetrically and independently of Chi, resulting in constitutive recombination activation. Genes Dev. 1999;13(7):901–911. | |
Anderson DG, Churchill JJ, Kowalczykowski SC. Chi-activated RecBCD enzyme possesses 5′–>3′ nucleolytic activity, but RecBC enzyme does not: evidence suggesting that the alteration induced by Chi is not simply ejection of the RecD subunit. Genes Cells. 1997;2(2):117–128. | |
Đermić D. Functions of multiple exonucleases are essential for cell viability, DNA repair and homologous recombination in recD mutants of Escherichia coli. Genetics. 2006;172(4):2057–2069. | |
Đermić D, Zahradka D, Petranović M. Exonuclease requirements for recombination of lambda-phage in recD mutants of Escherichia coli. Genetics. 2006;173(4):2399–2402. | |
Anderson DG, Churchill JJ, Kowalczykowski SC. A single mutation, RecB D1080A, eliminates RecA protein loading but not Chi recognition by RecBCD enzyme. J Biol Chem. 1999;274(38):27139–27144. | |
Jockovich ME, Myers RS. Nuclease activity is essential for RecBCD recombination in Escherichia coli. Mol Microbiol. 2001;41(4):949–962. | |
Ivanćić-Baće I, Peharec P, Moslavac S, Škrobot N, Salaj-Šmic E, Brćić-Kostić K. RecFOR function is required for DNA repair and recombination in a RecA loading-deficient recB mutant of Escherichia coli. Genetics. 2003;163(2):485–494. | |
Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432(7014):187–193. | |
Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol Cell. 2003;11(5):1337–1347. | |
Buljubašić M, Repar J, Zahradka K, Đermić D, Zahradka D. RecF recombination pathway in Escherichia coli cells lacking RecQ, UvrD and HelD helicases. DNA Repair (Amst). 2012;11(4):419–430. | |
Xu L, Marians KJ. PriA mediates DNA replication pathway choice at recombination intermediates. Mol Cell. 2003;11(3):817–826. | |
Wyatt HD, West SC. Holliday junction resolvases. Cold Spring Harb Perspect Biol. 2014;6(9):a023192. | |
Lesterlin C, Ball G, Schermelleh L, Sherratt DJ. RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature. 2014;506(7487):249–253. | |
Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464(7290):937–941. | |
Keyamura K, Sakaguchi C, Kubota Y, Niki H, Hishida T. RecA protein recruits structural maintenance of chromosomes (SMC)-like RecN protein to DNA double-strand breaks. J Biol Chem. 2013;288(41):29229–29237. | |
Harris DR, Pollock SV, Wood EA, et al. Directed evolution of ionizing radiation resistance in Escherichia coli. J Bacteriol. 2009;191(16):5240–5252. | |
Byrne RT, Klingele AJ, Cabot EL, et al. Evolution of extreme resistance to ionizing radiation via genetic adaptation of DNA repair. Elife. 2014;3: e01322. | |
Forget AL, Kowalczykowski SC. Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search. Nature. 2012;482(7385):423–427. | |
Xu HQ, Zhang AH, Auclair C, Xi XG. Simultaneously monitoring DNA binding and helicase-catalyzed DNA unwinding by fluorescence polarization. Nucleic Acids Res. 2003;31(14):e70. | |
Brody RS, Doherty KG, Zimmerman PD. Processivity and kinetics of the reaction of exonuclease I from Escherichia coli with polydeoxyribonucleotides. J Biol Chem. 1986;261(16):7136–7143. | |
Chase JW, Richardson CC. Exonuclease VII of Escherichia coli. Mechanism of action. J Biol Chem. 1974;249(14):4553–4561. | |
Viswanathan M, Lovett ST. Exonuclease X of Escherichia coli. A novel 3′–5′ DNase and Dnaq superfamily member involved in DNA repair. J Biol Chem. 1999;274(42):30094–30100. | |
Handa N, Morimatsu K, Lovett ST, Kowalczykowski SC. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes Dev. 2009;23(10):1234–1245. | |
Rath D, Mangoli SH, Pagedar AR, Jawali N. Involvement of pnp in survival of UV radiation in Escherichia coli K-12. Microbiology. 2012;158(Pt 5):1196–1205. | |
Viswanathan M, Dower KW, Lovett ST. Identification of a potent DNase activity associated with RNase T of Escherichia coli. J Biol Chem. 1998;273(52):35126–35131. | |
Ivanković S, Đermić D. DNA end resection controls the balance between homologous and illegitimate recombination in Escherichia coli. PLoS One. 2012;7(6):e39030. | |
Huertas P, Cortés-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455(7213):689–692. | |
Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284(14):9558–9565. | |
Coleman KA, Greenberg RA. The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem. 2011;286(15):13669–13680. | |
Barrangou R, Marraffini LA. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54(2):234–244. | |
Schiller CB, Seifert FU, Linke-Winnebeck C, Hopfner KP. Structural studies of DNA end detection and resection in homologous recombination. Cold Spring Harb Perspect Biol. 2014;6(10):a017962. | |
Cejka P, Cannavo E, Polaczek P, et al. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467(7311):112–116. | |
Niu H, Chung WH, Zhu Z, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467(7311):108–111. | |
Levikova M, Klaue D, Seidel R, Cejka P. Nuclease activity of Saccharomyces cerevisiae Dna2 inhibits its potent DNA helicase activity. Proc Natl Acad Sci U S A. 2013;110(22):E1992–E2001. | |
Harmon FG, Kowalczykowski SC. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 1998;12(8):1134–1144. | |
Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol Cell. 1999;3(5):611–620. | |
Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21(23):3085–3094. | |
Khakhar RR, Cobb JA, Bjergbaek L, Hickson ID, Gasser SM. RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol. 2003;13(9):493–501. | |
Lenhart JS, Brandes ER, Schroeder JW, Sorenson RJ, Showalter HD, Simmons LA. RecO and RecR are necessary for RecA loading in response to DNA damage and replication fork stress. J Bacteriol. 2014;196(15):2851–2860. | |
Cromie GA. Phylogenetic ubiquity and shuffling of the bacterial RecBCD and AddAB recombination complexes. J Bacteriol. 2009;191(16):5076–5084. | |
Capaldo FN, Barbour SD. DNA content, synthesis and integrity in dividing and non-dividing cells of rec- strains of Escherichia coli K12. J Mol Biol. 1975;91(1):53–66. | |
Zahradka K, Buljubašić M, Petranović M, Zahradka D. Roles of ExoI and SbcCD nucleases in “reckless” DNA degradation in recA mutants of Escherichia coli. J Bacteriol. 2009;191(5):1677–1687. | |
Repar J, Briški N, Buljubašić M, Zahradka K, Zahradka D. Exonuclease VII is involved in “reckless” DNA degradation in UV-irradiated Escherichia coli. Mutat Res. 2013;750(1–2):96–104. | |
Truong LN, Li Y, Shi LZ, et al. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci U S A. 2013;110(19):7720–7725. | |
Thoms B, Borchers I, Wackernagel W. Effects of single-strand DNases ExoI, RecJ, ExoVII, and SbcCD on homologous recombination of recBCD+ strains of Escherichia coli and roles of SbcB15 and XonA2 ExoI mutant enzymes. J Bacteriol. 2008;190(1):179–192. | |
Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273(34):21447–21450. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.