Back to Journals » Infection and Drug Resistance » Volume 13
Distribution of Genes Encoding Virulence Factors and the Genetic Diversity of Enteroinvasive Escherichia coli (EIEC) Isolates from Patients with Diarrhea in Ahvaz, Iran
Authors Farajzadeh-Sheikh A, Savari M, Ahmadi K , Hosseini Nave H, Shahin M, Afzali M
Received 17 October 2019
Accepted for publication 10 December 2019
Published 10 January 2020 Volume 2020:13 Pages 119—127
DOI https://doi.org/10.2147/IDR.S235009
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Suresh Antony
Ahmad Farajzadeh-Sheikh, 1, 2 Mohammad Savari, 1, 2 Khadijeh Ahmadi, 2, 3 Hossein Hosseini Nave, 4 Mojtaba Shahin, 5 Maryam Afzali 2
1Department of Microbiology, School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 2Infectious and Tropical Diseases Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 3Abadan Faculty of Medical Sciences, Abadan, Iran; 4Department of Microbiology and Virology, School of Medicine, Kerman University of Medical Sciences, Kerman, Iran; 5Department of Medical Laboratory Sciences, Faculty of Medical Sciences, Islamic Azad University, Arak, Iran
Correspondence: Maryam Afzali
Department of Microbiology, School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Golestan Blvd 39345-61355, Ahvaz, Iran
Tel +09156127753
Fax +98-61-3333 2036
Email [email protected]
Background: Entero-invasive E. coli (EIEC) is one of the causes of bacillary dysentery in adults and children. The ability of EIEC to invade and colonize the surface of epithelial cells is influenced by many virulence factors. This study aimed to investigate the distribution of virulence factor genes in EIEC strains isolated from patients with diarrhea in Ahvaz, Iran, as well as the genetic diversity between these isolates by Multilocus variable-number tandem repeat analysis (MLVA).
Materials and Methods: A total of 581 diarrheic stool samples were collected from patients with diarrhea attending two hospitals, in Ahvaz, Iran. The E. coli strains were identified by biochemical methods. Subsequently, all E. coli isolates were identified as EIEC by polymerase chain reaction (PCR) for the ipaH gene. The EIEC isolates evaluated by PCR for the presence of 8 virulence genes (ial, sen, virF, invE, sat, sigA, pic, and sepA). All EIEC strains were genotyped by the MLVA typing method.
Results: A total of 13 EIEC isolates were identified. The presence of ial, virF, invE, sen, sigA, pic, and sat genes was confirmed among 92.3%, 84.6%, 84.6%, 76.9%, 69.2%, and 15.3% of EIEC isolates, respectively. On the other hand, none of the isolates were positive for the sepA gene. The EIEC isolates were divided into 11 MLVA types.
Conclusion: Our results showed a high distribution of virulence genes among EIEC isolates in our region. This study showed that MLVA is a promising typing technique for epidemiological studies. MLVA can supply data in the form of codes that can be saved in the database and easily shared among laboratories, research institutes, and even hospitals.
Keywords: entero-invasive Escherichia coli, diarrhea, virulence factor, MLVA
Introduction
Entero-invasive Escherichia coli (EIEC) is one of the pathotypes of diarrheagenic Escherichia coli (DEC) that cause shigellosis-like symptoms in both children and adults.1 Although EIEC infections occur worldwide, these are particularly common in low-income countries with poor hygiene.2 EIEC, like Shigella, is responsible for bacillary dysentery. Bacillary dysentery is characterized by fever, abdominal cramps, diarrhea, sometimes vomiting, and the stool contains blood and pus.3 EIEC can cause invasion and penetration of the epithelial cells of the colon. After the penetration into the colonic mucosa, EIEC proliferates intracellularly and spread to adjacent cells and destroying the intestinal epithelial barrier.2 The genes related to the invasion of EIEC are located on a large virulence inv plasmid and chromosome. The inv plasmid is an essential virulence determinant of EIEC which encodes the type III secretion system necessary for attachment, invasion of the host cell and intercellular spread.4 Clinical phenotypes are determined by different virulence genes and host immune system activity. They are often multifactorial and coordinately regulated, and the genes tend to be clustered in the genome.5 The epithelial cell penetration and modification of the host response for dissemination from cell to cell are mediated by an invasion-associated locus (ial), which is located on a plasmid and the invasion plasmid antigen H (ipaH) genes present in both chromosome and plasmid. Because the ipaH gene is present as multiple copies on the inv plasmid and the chromosome, it is used as a diagnostic marker for EIEC detection.6,7 virF and invE are two regulatory proteins that control the transcriptional of invasion genes.7 The serin autotransporters of proteins Enterobacteriaceae (SPATEs) are present in EIEC strains. The SPATEs family has been classified into 2 classes. Class 1 SPATEs are toxic for epithelial cells, include the Shigella IgA-like protease homolog gene (sigA) and secreted autotransporter toxin gene (sat) which are directly cytotoxic for epithelial cells. The sat gene is located in the chromosome and promotes serine protease activity and displays cytopathic activity in several intestinal cell lines. Class 2 SPATEs members, including the protease involved in colonization of the intestine (pic) and the extracellular protein Shigella A (sepA), contribute to intestinal inflammation and colonization.8 Two enterotoxins, Shigella enterotoxin 1 (ShET1) and Shigella enterotoxin 2 (ShET2), transport water and electrolyte in the small intestine and cause diarrhea. ShET-2 which encoded by the sen gene is found commonly among EIEC strains and believed to be involved in the invasion process.9 EIEC isolates harboring these virulence genes that can induce the inflammation and extensive mucosal damages in intestinal infections, especially when these strains encode more than one of the virulence factors. Therefore, understanding the distribution of virulence genes in EIEC isolates could be useful for researchers designing new antibiotics against them. Virulence therapies do not damage the normal flora of the host, because this therapy targets virulence pathways that do not exist in the nonpathogenic bacteria. For example, because of the importance of virF in regulating viral genes, new antibiotics targeting the virF gene have received much attention.10 Furthermore, the molecular epidemiology of such isolates is very helpful. Multilocus variable-number tandem repeat analysis (MLVA) is an effective molecular typing method based on counting the number of Variable Number Tandem Repeat (VNTR) loci. In recent years, MLVA has been applied to investigate the clonal relationships among isolates of E. coli strains.11 Despite many reports about the prevalence and antimicrobial resistance of EIEC isolates from different parts of the world including Iran, investigations about the occurrence of virulence factors are still rare worldwide. Previous studies showed that EIEC is not endemic in Iran and our region.7,12,13 There is no information about the genetic diversity of EIEC strains in the southwest of Iran. Therefore, this study aimed to investigate the distribution of genes encoding virulence factors and the genetic diversity of EIEC strains isolated from patients with diarrhea by MLVA.
Materials and Methods
Ethics Statement
The study design was approved by the Research Ethics Committee of Ahvaz Jundishapur University of Medical Sciences, Iran (IR.AJUMS.REC.1396.434). As a part of the Ahvaz Jundishapur University of Medical Sciences policy, written informed consent was obtained from all of the adult patients and children’s parents or legal guardians of any patient under the age of 18 years. The study was conducted in accordance with the Declaration of Helsinki.
The Collection of Samples
In this cross-sectional study, 581 stool specimens were collected from patients with diarrhea referring to the teaching hospitals (Golestan and Abouzar hospitals), in Ahvaz, southwest of Iran, from September 2016 to August 2017. Patients with a history of fever, nausea, vomiting, abdominal cramps, watery, mucoidal and bloody diarrhea were included in our study. Also, they had not used the antibiotic nearly 2 weeks before. If patients had taken antibiotics in the last two weeks, they would be excluded from our study. Stool samples initially were cultured on MacConkey agar (Merck, Germany), and incubated for 24 hrs at 37°C. The lactose positive colonies were tested by standard biochemical and bacteriological tests such as Triple Sugar Iron Agar, Indole test, Methyl red and Voges-Proskauer tests, and Simmons citrate agar (Merck, Germany) for detection of E. coli isolates.13 All isolates that confirmed as E. coli, were preserved in Tryptic Soy Broth (TSB) (Merck, Germany), containing glycerol (30%) at −70°C.
Molecular Confirmation of EIEC Strains
All E. coli isolates were evaluated for the presence of the ipaH gene by PCR. DNA extraction of E. coli isolates was performed by the boiling method previously described.14 The sequences of primers used for the detection of the ipaH gene are shown in Table 1. The reaction volume was performed in a final volume of 25 μL containing 1X PCR buffer, 1U Taq polymerase, 1 μM MgCl2, 200 μM of dNTPs (CinnaGen, Iran), 0.2 μL of each primer, and 3 μL of DNA template. The amplification reaction was programmed by a thermal cycler (Eppendorf, Germany) as follows: Initial denaturation at 94°C for 5 min, 35 cycles of 94°C for 30 s, annealing 58°C for 45 s, extension 72°C for 1min and final extension 72°C for 5 min. The PCR products were separated on a 1.5% agarose gel containing ethidium bromide and finally visualized in the gel documentation system (Protein simple, USA). Shigella flexnery ATCC 12122 was used as a positive PCR control for the ipaH gene.
 |
Table 1 Primers Used to Identify Virulence-Associated Genes of EIEC |
Amplification of Virulence Factor Genes
PCR was carried out on all EIEC strains to evaluate the prevalence of the ial, virF, invE, sen, sat, sigA, pic, and sepA genes.8,9 The sequences of primers and annealing temperatures of virulence factor genes are shown in Table 1. The total volume of the PCR mixture was 25 µL, containing 0.5 µL of DNA template, 1X PCR buffer, 2.5 Mm of MgCl2, 0.5 µL each virulence gene primer, 0.5 µL Taq DNA polymerase. The PCR conditions for the amplification of virulence genes included an initial denaturation at 94°C for 60 seconds, 35 cycles of denaturation at 94°C for 60 seconds, annealing (variable) for 60 seconds, and extension at 72°C for 60 seconds, as well as a final extension at 72°C for 7 mins. After performing PCR, the size of each locus was easily determined on 1.5% gel agarose. Positive controls for each gene were as follows: S. flexnery ATCC 12122 for virF, EIEC strain 44825 for invE, EIEC strain 43893 for ial, S. flexneri 2a strain 2457T is for sat, sepA, pic and sigA, S. flexnery 4a strain 12023 for sen.
Genotyping of EIEC Isolates by MLVA Analysis
MLVA was performed for all EIEC isolates. Seven VNTR loci were selected for the genetic typing of the EIEC isolates. PCR was carried out as described by a previous study.15 The primers and repeat sizes for each locus are shown in Table 2. The PCR products were electrophoresed on a 1.5% agarose gel containing ethidium bromide and visualized in a gel documentation system (Protein simple, USA). The copy numbers of the repeat for each isolate was calculated by the following formula:
 |
Table 2 Locus-Specific PCR Primers Selected for MLVA |
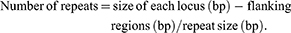
The results were imported into Microsoft Excel 2010 software and analyzed with the Bionumerics Software v.6.6 (Applied maths, Sint-Martens-Latem, Belgium). For clustering, a cut-off value of 90% similarity was used. The dendrogram of genetic relationships was generated using the Unweighted Pair Group Method with Average linkages (UPGMA).16
Statistical Analysis
The results were analyzed, using SPSS software version 22 to obtain frequencies and comparisons among clones. A nonparametric Chi-square test was used and a P-value of < 0.05 was considered statistically significant.
Results
Study Population
Of 581 fecal specimens 43.2% (n=251), were DEC strains confirmed by standard biochemical and microbiological tests. The patient’s age range was between 0 to 81 years.
Distribution of EIEC
Out of 251 E. coli isolates, 5.1% (n=13) were positive for ipaH. All the EIEC strains have been isolated from children. Of 13 strains of EIEC, 84.6% (n=11) isolated from children under the age of 2 years (P<0.05). Children were categorized into five different groups according to their age: (0–12), (13–24), (25–36), (35–48), and (49–60) months. Distribution of EIEC strains according to age, gender, and clinical symptoms are shown in Table 3.
 |
Table 3 Distribution EIEC Strains According to Seasons of Sampling, Age, Gender and Clinical Symptoms |
Prevalence of Virulence Factors Genes Among EIEC Strains
The detection of the virulence genes from 13 EIEC isolates showed that 92.3% (n=12) of isolates were positive for ial, whereas 84.6% (n=11) were positive for the invE and virF genes. The data revealed that 76.9% (n==10), 69.2% (n=9), 30.7% (n=4), and 15.3% (n=2) of strains were positive for the sen, sigA, pic, and sat genes, respectively. None of the isolates were positive for the sepA gene. There were six distinct virulence patterns in our isolates. The most prevalent Virulence patterns were, ipaH+, ial+, virF+, invE+, sen+, sigA+ found in 30.7% (n=4) strains, followed by ipaH+, ial+, virF+, invE+, sen+ 23.1% (n=3). The virulence patterns of all EIEC isolates are shown in Figure 1.
 |
Figure 1 UPGMA dendrogram based on MLVA type of isolates in regard to their virulence gene. +: Present, -: Absent. |
MLVA Assay
MLVA performed for all EIEC isolates. Analysis of the MLVA profiles using UPGMA showed that all EIEC isolates were grouped into 11 distinct MLVA types with 2 clusters and 9 singletons, and 2 multitone genotypes. The six virulence patterns were showed with their MLVA pattern in Figure 1. Some EIEC isolates with the same MLVA type (M8 and M10) had different virulence patterns. Minimal spanning tree (MST) of virulence genes distribution among different MLVA patterns is shown in Figure 2. Each circle denotes an MLVA type, with the number of isolates in each type, as indicated within the circle.
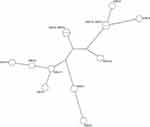 |
Figure 2 Spanning tree of virulence genes distribution among different MLVA patterns. |
Discussion
EIEC is one of the most important E. coli pathotypes that cause watery diarrhea and dysentery. Unfortunately, there are limited reports on the prevalence of EIEC in Iran. Due to the inability of conventional culture methods to detect pathogenic E. coli from non-pathogenic isolates, EIEC is usually ignored.13 In the current study, the ipaH gene was used for detecting of EIEC isolates. The invasive genes ipaH was positive for all (100%) the isolates which agree with various other studies done so far.7,13 The ipaH gene is more stable because it presents multiple copies on both chromosomes and the inv plasmid and seemed to be less compromised by plasmid loss and/or deletions. In our study, the prevalence of EIEC among DEC isolates was (5.1%) by PCR. This was in agreement with the findings in the previous study from Shiraz (5.5%).17 Compared with our findings, lower prevalence rates of EIEC have been reported from Northern Iran (0.5%),18 Nigeria and Vietnam (0.8%).19,20 The high frequencies of the ipaH gene could explain frequencies of fever, vomiting, and dehydration in infected children. The EIEC isolates are similar to Shigella in the pathogenesis. Several virulence factors associated with EIEC pathogenesis have been identified. However, very little research has been done to identify their virulence factors profiles. This study, for the first time in Ahvaz, investigated the prevalence and distribution of virulence genes in EIEC isolates. The severity of the disease depends on the virulence factors of the bacterial strains. Among virulence factors in EIEC isolates, the ial gene which is located on a large plasmid facilitates the penetration of this bacterium into epithelial cell.21 In the current study, (92.3%) of isolates were positive for the ial gene. In research conducted by Hosseini Nave et al all isolates were positive for the ial gene.7 The ial gene cluster is located near a region of the plasmid, which is a hot spot for spontaneous deletions. This probably explains the lower prevalence of ial than ipaH in the EIEC strains. The expression of virulence genes is regulated by the heat-stable nucleotide structural protein (H-NS), which regulates their transcription for invasion. Transcription starts from the virF gene, which then turns on the transcription of the invE regulatory genes. Subsequently, the invE protein reverses the H-NS induced inhibition of transcription, which eventually elicits virulence genes on the plasmid.3,22 In our study, both virF and invE were found in (84.6%) of EIEC isolates. These results are approximately consistent with the previous study.7 Since virF and invE genes are located on the virulence plasmid, they are susceptible to deletion. This may be the reason for the reduced frequency of virF and invE among the isolates. A new study has shown the use of regulatory genes like virF as a target for novel antibiotics.10 ShET-1 and ShET-2 play an important role in stimulating fluid secretion into the small intestine, thus, contributing to the watery phase of diarrhea. ShET-2 is encoded by the sen gene. This gene is located on a plasmid and originally discovered in EIEC isolates. Studies have shown that ShET-2 contributes to epithelial inflammation and diarrhea.9,23 In our study, the sen gene was found in (76.9%) of EIEC strains. Therefore, it can be predicted that sen is the major player to cause the symptoms of electrolyte imbalance and water loss causing dehydration among EIEC strains in our region. The SPATE genes are virulence factors that secreted autotransporters in gram-negative bacteria. There is very little information about the distribution of SPATE genes in EIEC isolates. In the present study, the sigA gene was recognized as the most common SPATE gene among EIEC isolates. In agreement with our study, Hosseini Nave et al and Boisen et al found the high rates of this gene among EIEC isolates.7,8 These results implied that sigA may play an important role in the pathogenesis of EIEC. The pic gene was detected in 30.7% of the EIEC isolates. The frequency of this gene was relatively similar to the previous study from Kerman, Iran.7 In this study, all isolates were negative for the SepA. Our results matched with the findings of studies conducted by Hosseini Nave et al and Boisen et al8 The proteases are encoded by the sepA gene located in the virulence plasmid, and by the sigA and pic genes located in the chromosome. Due to storage/subculturing, the plasmid might have been lost together with the sepA gene. The results of the distribution of the sat gene in the present study showed that (15.3%) of EIEC isolates were positive for this gene which is different from prior.7,8
Molecular typing methods are used to determine genetic relationships between pathogenic strains for epidemiological surveillance. MLVA can provide data in the form of codes that can be saved in the database and transferable between different laboratories. This method, compared with other typing methods, such as pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) is a rapid and low-cost genotyping method that has differentiated closely related strains of bacteria from each other.11,24 The MLVA also allowed us to establish associations between genotypes and parameters such as virulence characteristics.25 In this study, we used the MLVA method for the typing of EIEC isolates. We selected VNTR loci which can produce the most discriminatory power for E. coli and easily analyzed on the agarose gel. Therefore, this method can be performed for epidemiological purposes in a laboratory with simple molecular tools. The UPGMA clustering of the MLVA data showed that there were 2 major clusters with different 11 MLVA types. In agreement with our study, Hosseini Nave et al showed that EIEC isolates belonged to two clonal complexes.7 The current study findings demonstrated heterogeneity among EIEC isolates and it seemed to be due to the horizontal spread of mobile genetic elements. In the current investigation, some EIEC isolates with the same MLVA type had different virulence gene patterns. This might be because many virulence genes only located on the virulent plasmid that is susceptible to deletion and loss during storage and subculturing. Understanding these patterns of EIEC infection and transmission would provide important information on how best to design intervention and control strategies targeted at EIEC.
Conclusion
The present study provided insights into some baseline information about the distribution of some virulence genes of EIEC isolates in Ahvaz Province in Iran. This study indicated that epidemiological programs are necessary to monitor the distribution of virulence genes locally for the prevention of the spread of the EIEC isolates harboring them. As mentioned above, MLVA typing is a much easier and more rapid technique for the analysis of E. coli strains relatedness. This study showed that MLVA is a promising typing technique for epidemiological studies. MLVA can supply data in the form of codes that can be saved in the database and easily shared among laboratories, research institutes, and even hospitals.
Acknowledgments
The authors would like to thank the Infectious and Tropical Diseases Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Iran for financial support.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Pereira A, Giugliano L. Adhesion of diarrheagenic Escherichia coli and inhibition by glycocompounds engaged in the mucosal innate immunity. Biology. 2013;2(2):810–831. doi:10.3390/biology2020810
2. Pasqua M, Michelacci V, Di Martino ML, et al. The intriguing evolutionary journey of enteroinvasive E. coli (EIEC) toward pathogenicity. Front Microbiol. 2017;8:2390. doi:10.3389/fmicb.2017.02390
3. Mohammadzadeh M, Goudarzi H, Dabiri H, Fallah F. Molecular detection of lactose fermenting enteroinvasive Escherichia coli from patients with diarrhea in Tehran-Iran. Iran J Microbiol. 2015;7(4):198.
4. Cowley LA, Oresegun DR, Chattaway MA, Dallman TJ, Jenkins C. Phylogenetic comparison of enteroinvasive Escherichia coli isolated from cases of diarrhoeal disease in England, 2005–2016. JMM. 2018;67(6):884–888. doi:10.1099/jmm.0.000739
5. Peng J, Yang J, Jin Q. The molecular evolutionary history of Shigella spp. and enteroinvasive Escherichia coli. Infect Genet Evol. 2009;9(1):147–152. doi:10.1016/j.meegid.2008.10.003
6. Song T, Toma C, Nakasone N, Iwanaga M. Sensitive and rapid detection of Shigella and enteroinvasive Escherichia coli by a loop-mediated isothermal amplification method. FEMS Microbiol Lett. 2005;243(1):259–263. doi:10.1016/j.femsle.2004.12.014
7. Hosseini Nave H, Mansouri S, Moghadam MT, Moradi M. Virulence gene profile and multilocus variable-number tandem-repeat analysis (MLVA) of enteroinvasive Escherichia coli (EIEC) isolates from patients with diarrhea in Kerman, Iran. Jundishapur J Microbiol. 2016;9:6. doi:10.5812/jjm.33529
8. Boisen N, Ruiz-Perez F, Scheutz F, Krogfelt KA, Nataro JP. High prevalence of serine protease autotransporter cytotoxins among strains of enteroaggregative Escherichia coli. Am J Trop Med Hyg. 2009;80(2):294–301. doi:10.4269/ajtmh.2009.80.294
9. Cruz C, Souza M, Serra PT, et al. Virulence factors associated with pediatric shigellosis in Brazilian Amazon. Biomed Res Int. 2014;2014. doi:10.1155/2014/539697
10. Emanuele AA, Adams NE, Chen Y-C, Maurelli AT, Garcia GA. Potential novel antibiotics from HTS targeting the virulence-regulating transcription factor, VirF, from Shigella flexneri. J Antibiot (Tokyo). 2014;67(5):379. doi:10.1038/ja.2014.10
11. Ranjbar R, Memariani M. Multilocus variable-number tandem-repeat analysis for genotyping of Shigella sonnei strains isolated from pediatric patients. Gastroenterol Hepatol Bed Bench. 2015;8(3):225.
12. Broujerdi SM, Ardakani MR, Rezatofighi SE. Characterization of diarrheagenic Escherichia coli strains associated with diarrhea in children, Khouzestan, Iran. J Infect Dev Ctries. 2018;12(08):649–656. doi:10.3855/jidc.9538
13. Abbasi P, Kargar M, Doosti A, Mardaneh J, Dalini SG, Dehyadegari MA. Real time pcr for characterization of enteroinvasive E. coli (eiec) in children with diarrhea in shiraz. Ann Colorectal Res. 2014;2(3). doi:10.17795/acr-2272
14. Memariani M, Peerayeh SN, Salehi TZ, Mostafavi SKS. Occurrence of SHV, TEM and CTX-M β-lactamase genes among enteropathogenic Escherichia coli strains isolated from children with diarrhea. Jundishapur J Microbiol. 2015;8:4. doi:10.5812/jjm.8(4)2015.15620
15. Gorgé O, Lopez S, Hilaire V, Lisanti O, Ramisse V, Vergnaud G. Selection and validation of a multilocus variable-number tandem-repeat analysis panel for typing Shigella spp. JCM. 2008;46(3):1026–1036. doi:10.1128/JCM.02027-07
16. Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. MIRU-VNTR plus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38(suppl_2):W326–W331. doi:10.1128/JCM.02027-07
17. Aminshahidi M, Arastehfar A, Pouladfar G, Arman E, Fani F. Diarrheagenic Escherichia coli and Shigella with high rate of extended-spectrum Beta-lactamase production: two predominant etiological agents of acute diarrhea in Shiraz, Iran. Microb Drug Resist. 2017;23(8):1037–1044. doi:10.1089/mdr.2017.0204
18. Moshtagian F, Alipour M, Yahyapour Y. Prevalence of Escherichia coli pathotypes among children with diarrhea in Babol, Northern Iran. Int J Enteric Pathog. 2016;4(3):1–4. doi:10.15171/ijep.2016.01
19. Ifeanyi CIC, Ikeneche NF, Bassey BE, Al-Gallas N, Aissa RB, Boudabous A. Diarrheagenic Escherichia coli pathotypes isolated from children with diarrhea in the Federal Capital Territory Abuja, Nigeria. J Infect Dev Ctries. 2015;9(02):165–174. doi:doi.10.3855/jidc.5528
20. Hien BTT, Scheutz F, Cam PD, et al. Diarrheagenic Escherichia coli and Shigella strains isolated from children in a hospital case-control study in Hanoi, Vietnam. JCM. 2008;46(3):996–1004. doi:10.1128/JCM.01219-07
21. Sousa MÂB, Mendes EN, Collares GB, Péret-Filho LA, Penna FJ, Magalhães PP. Shigella in Brazilian children with acute diarrhoea: prevalence, antimicrobial resistance and virulence genes. Mem Inst Oswaldo Cruz. 2013;108(1):30–35. doi:10.1590/S0074-02762013000100005
22. Fan W, Qian H, Shang W, et al. Low distribution of genes encoding virulence factors in Shigella flexneri serotypes 1b clinical isolates from eastern Chinese populations. Gut Pathog. 2017;9(1):76. doi:10.1186/s13099-017-0222-9
23. Roy S, Thanasekaran K, Dutta Roy AR, Sehgal SC. Distribution of Shigella enterotoxin genes and secreted autotransporter toxin gene among diverse species and serotypes of Shigella isolated from Andaman Islands, India. Trop Med Int Health. 2006;11(11):1694–1698. doi:10.1111/j.1365-3156.2006.01723.x
24. Naseer U, Olsson-Liljequist BE, Woodford N, et al. Multi-locus variable number of tandem repeat analysis for rapid and accurate typing of virulent multidrug resistant Escherichia coli clones. PLoS ONE. 2012;7(7):e41232. doi:10.1371/journal.pone.0041232
25. Bustamante AV, Sanso AM, Parma AE, Lucchesi PMA. Subtyping of STEC by MLVA in Argentina. Front Cell Infect Microbiol. 2012;2:111. doi:10.3389/fcimb.2012.00111
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
