Back to Journals » International Journal of General Medicine » Volume 15
Discovering Novel Biomarkers Associated with the Pathogenesis of Psoriasis: Evidence from Bioinformatic Analysis
Authors Yang Y , Xie S, Jiang W, Tang S, Shi Y
Received 12 January 2022
Accepted for publication 1 March 2022
Published 11 March 2022 Volume 2022:15 Pages 2817—2833
DOI https://doi.org/10.2147/IJGM.S354985
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Yang Yang,1,2 Shaoqiong Xie,1 Wencheng Jiang,1 Suwei Tang,1,2 Yuling Shi1,2
1Department of Dermatology, Shanghai Skin Disease Hospital, School of Medicine, Tongji University, Shanghai, 200443, People’s Republic of China; 2Institute of Psoriasis, School of Medicine, Tongji University, Shanghai, 200092, People’s Republic of China
Correspondence: Yuling Shi; Suwei Tang, Department of Dermatology, Shanghai Skin Disease Hospital, Tongji University School of Medicine, 1278 Baode Road, Jing-an District, Shanghai, 200443, People’s Republic of China, Tel +86-13816213884 ; +86-18017337631, Fax +63833136, Email [email protected]; [email protected]
Objective: This study aimed to investigate key biomarkers and their molecular pathogenesis in psoriasis.
Methods: Differentially expressed genes (DEGs) of datasets (GSE13355, GSE30999, and GSE106992) obtained from Gene Expression Omnibus (GEO) were identified using Venn diagram. Function and pathway enrichment analyses were performed. Protein–protein interaction (PPI) network and the hub genes were constructed. The correlation between normal tissue and infiltrating immune cells was analyzed by CIBERSORT. ROC analysis was performed to distinguish between skin lesion samples and skin non-lesion samples. Analyze the highest expression of single gene in the whole body within the Human Protein Atlas (HPA) database. Effect of CXCL8 expression level on proliferation, invasion, migration and apoptosis of HaCat cells was detected by qPCR.
Results: A total of 239 pairs of normal and lesional skin samples were downloaded. PPI network revealed a tight interaction among 197 DEGs. The GO enrichment analysis showed that these genes were markedly enriched in the “defense response to virus”, “type I interferon signaling pathway”, and “cell response to type I interferon” categories. The KEGG pathway analysis showed that the DEGs were mainly in the NOD-like receptor axis, interaction between cytokine and cytokine receptor and the IL− 17 axis. PPI analysis showed that CXCL8 was the novel hub gene of psoriasis and correlated to 22 types of infiltrating immune cells. 6 miRNAs were predicted to be related to CXCL8. CXCL8 was most widely distributed in lymphoid tissues and plays a role in psoriatic inflammatory lesions by promoting cell proliferation, migration, and anti-apoptosis.
Conclusion: CXCL8 plays a key role in psoriasis development. This study provided new insights into the exploration of molecular mechanisms and therapeutic targets of psoriasis.
Keywords: psoriasis, CXCL8, molecular mechanism, bioinformatics
Introduction
Psoriasis is a chronic inflammatory skin Disease that negatively affects the quality of life by significantly impairing physical and psycho-social health.1 Psoriasis is also known as a systemic disease accompanied by multiple comorbidities.2 People with psoriasis are more frequently affected by arterial hypertension, diabetes mellitus, coronary artery disease, dyslipidemia, obesity, and metabolic syndrome.3–5 The multifactorial relation between psoriasis and obesity leads to acceleration of systemic inflammation, intensification of skin lesions, decreased activity, stigmatization, and poor eating habits or psychological disorders. Fortunately, with better understanding of psoriasis,6 an increasing number of therapies have emerged. The mainstream psoriatic therapy includes systemic application of retinoic acid, immunosuppressant, biological agents, external use of glucocorticoid, retinoic acid drugs, and so on. However, the adverse reactions are serious. Due to the unclear etiology and complex pathogenesis of psoriasis, it still remains incurable. Hence, there is great urgency in the comprehension of molecular networks involved in psoriasis to enhance the quality of psoriatic care.
Psoriasis is mediated by polygenic inheritance T cells. The excessive activation of immune cells is caused by a variety of pathogenic factors. At present, multiple new drugs are available for the management of psoriasis. The biological agents used for psoriasis therapy in China are anti-TNF-α, anti-IL17A, anti-IL12/23, and anti-IL23. Anti-TNF-αwas the first commercially available biological agent for psoriasis, and it includes Etanercept, Adalimumab, and Infliximab. Among them, Etanercept sequesters serum TNF-α, which limits its activity, and produces an anti-inflammatory response.7 Likewise, Adalimumab also binds to TNF - α and blocks its interaction with the TNF receptor on the surface of p55 and p75 cells, thereby reducing inflammation. Lastly, Infliximab is a human mouse chimeric monoclonal antibody, which strongly sequesters both soluble and transmembrane forms of TNF - α, thereby making them inactive.8 According to reports, Adalimumab and Infliximab fared better than Etanercept in controlling psoriatic skin lesions. Moreover, the onset speed of Adalimumab and Infliximab remained slower than that of anti-IL17A.9 The anti-IL17A drugs prevalent in China are Scuchizumab and Iqizumab. Both drugs have similar efficacy in psoriatic lesion resolution and have a better rate of improvement in PASI than monoclonal antibodies.10–12 Despite the presence of multiple psoriatic drugs, there is still potential for advancements, especially in terms of making the drugs affordable and in reducing double infection risk. Hence, understanding the intricate details of psoriatic pathogenesis will add value to the future of psoriatic care.
The CXC family plays an important role in mediating pathogenesis of psoriasis.13 Some treatment methods for psoriasis may exert therapeutic effects by affecting the expression of chemokines and receptors, and may be related to the severity of psoriasis.14 Some chemokines stimulate the chemotactic induction of inflammatory T cells (such as CXCL10, CXCL16) and neutrophils (such as CXCL1 and CXCL5) during psoriasis. CXCL8 is known to regulate neutrophil infiltration, angiogenesis, and keratinocyte proliferation in psoriatic lesions. It is worth mentioning that serum CXCL8 in psoriatic patients remains significantly elevated and is positively correlated with the severity of the disease.15,16 Nevertheless, the specific function of CXCL8 in psoriatic progression remains unknown and requires further investigation.
Over the past decade, advances in microarray screening and bioinformatics investigations have enabled the genome-wide search for differentially expressed genes (DEGs) and physiological networks linked to specific diseases, including psoriasis. Yet, there have been limitations. For example, a high false-positive incidence was noted in an isolated microarray study. Additionally, differences in microarray procedures, variations in statistical analysis, and limited sample populations may have, in unison, introduced discrepancies in conclusions across studies. Hence, additional extensive examinations are warranted to reliably recognize psoriasis-related DEGs in the search of effective targeted therapy and prognostic biomarkers for psoriasis.
The goal of our work was to analyze DEGs, collected from three microarray datasets from the Gene Expression Omnibus (GEO) database, comparing psoriatic skin to healthy skin. Additionally, we conducted network analyses to further evaluate underlying molecular mechanisms involved in psoriasis. Using the results of our analysis, we established an extensive network of genes and functional pathways involved in psoriasis. Moreover, we demonstrated CXCL8 to be a promising psoriatic biomarker. Effect of CXCL8 expression level on proliferation, invasion, migration and apoptosis of HaCat cells were detected by qPCR.
Materials and Methods
Data Collection and Preprocessing
The raw psoriatic gene expression data (cel files), including GSE13355 (58 psoriatic skin specimen verses 64 healthy skin specimen), GSE30999 (82 psoriatic skin specimen verses 82 healthy skin specimens), and GSE106992 (125 psoriatic skin specimen verses 67 healthy skin specimen), were retrieved from the GEO database (https://www.ncbi.nlm.nih.gov).17 Simultaneously, the hall marker gene information was obtained from the Molecular Signatures Database (MSigDB) (https://www.gsea-msigdb.org/gsea/msigdb).18 Lastly, the gene expression data of 33 cancer types were obtained from the UCSC Xena (http://xena.ucsc.edu/).19 The study was approved by the Ethics Review Committee of Shanghai Skin Disease Hospital.
Screening of DEGs
DEG analysis was done with R 3.4.1ʹs (https://www.r-project.org/) Affy and Limma package.20,21 The corresponding plots were made by R and the genes that followed the following criteria were regarded as DEGs: P < 0.05 and log |FC|>1.0. Finally, we generated Venn charts,22 including an overlap of total, upregulated, and downregulated DEGs from all 3 datasets.
GO and KEGG Enrichment Analyses of DEGs
Gene Ontology (GO)23 enrichment analysis and Kyoto encyclopedia of Genes and Genomes (KEGG)24 functional network analysis were conducted using “clusterProfiler”25 within R. This allowed us to recognize potential DEG roles and relationships with corresponding diseases. Gene sets carrying a p value <0.05 was regarded as significant enrichment.
GSEA Analysis
The relationship between CXCL8 and functional network genes was analyzed with gene set enrichment analysis (GSEA, v2.2,http://www.broad.mit.edu/gsea/).26 Furthermore, its association with phenotype was explored with TCGA gene sets that included 33 forms of cancer from MSigDB. The gene sets that showed marked enrichment by genes, based on elevated CXCL8levels, [false discovery rate (FDR) <0.05] were defined as enriched gene sets.
Integration of the Protein–Protein Interaction (PPI) Network
The PPI network was estimated with the online Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org)(version 10.0)27 database and it was formed with the STRING database, with a combined interaction score >0.4 deemed as significant. Cytoscape (version 3.4.0) was employed for the visualization of potential underlying signaling pathways.28 Finally, Ggplot2 package29 was used to generate a bar chart illustrating distribution of relevant genes.
Immune Infiltration Analysis
CIBERSORT was employed for immune cell infiltration analysis30 and facilitated the identification of alterations in infiltrating immune cell between psoriatic and healthy skin specimen. The corresponding bubble graph was visualized with ggplot2 package.
Receiver Operating Characteristic (ROC) Curve Analysis
ROC curve analysis, conducted with the “pROC” package of R,31 was chosen for determining specificity and sensitivity of the diagnosis.32 The maximum value of the sum of specificity and sensitivity was set as the threshold for individual hub genes.
Construction of the ceRNA Network
The relationship between DElncRNAs and DEmiRNAs were estimated with miRcode.33 The DEmiRNA-targeted mRNAs were obtained based on starBase,34 miRDB,35 and miRWalk.36 To be deemed as candidate mRNAs, the mRNAs had to be identified by all data sets and required intersection with DEmRNAs to remove DEmiRNAs-targeted DEmRNAs. Next the lncRNA–miRNA–mRNA ceRNA axis was generated using the DEmiRNA–DElncRNA and DEmiRNA–DEmRNA relationships, and was visualized with the Cytoscape software.28 Potential associations between ceRNA levels were analyzed with linear regression.
Immunohistochemical (HIC) Analysis of Lymph Nodes Through HPA Database
To explore the diagnostic significance of CXCL8, whole-body distribution of this gene (both at the transcript and protein level) was examined using the Human Protein Atlas (HPA, https://www.proteinatlas.org/), an open access website providing protein maps within organs, tissues, and cells.37
Cell Transfection
Add 80 pmol of CXCL8-siRNA or NC-siRNA (no significant sequence) to 120 ul of Opti-MEM medium and 12 ul of HilyMax transfection reagent respectively and mix well. Let stand at room temperature for 15 min and then add into HaCaT (human skin keratin-forming cells, Purchased from GeneFull Biotech co.,Ltd). 37°C, 5% CO2 incubate for 6h and then change to fresh complete medium. RNA was extracted and the interference effect was verified using qPCR.
CCK-8 Assay
Cells were inoculated at a density of 1×104 cells per well in 96-well cell culture plates overnight at 37°. After discarding the solution, 100ul of DMEM medium was added to each well. Discard the old fluid at 0h, 24h, 48h and 72h. Add 95 ul of medium and 5 ul of CCK-8, incubate at 37°C for 1–3 h. Determine the OD value of each well at 450 nm.
Apoptosis Detection and Flow Cytometry
Cells were inoculated at a density of 1×105 cells per well in 24-well culture plates for 48 h. Cells were collected by digestion with 0.25% trypsin (without EDTA). Cells were washed twice with PBS and then once with Binding Buffer. Add Binding Buffer to resuspend the cells and adjust the cell concentration to 1×106 cells/mL. Add 5 μL of Annexin V-FITC and LPropidium Iodide to 100 ul of cell suspension and mix well. The reaction was carried out at room temperature and protected from light for 15–20 min, and then adjusted to 500ul by adding Binding Buffer. Cell apoptosis was detected by flow cytometry.
Transwell Invasion Assay
Using Matrigel plates containing serum-free RPMI 1640 medium, 200 ul of cell suspension was added to each well in the upper chamber, and 500 ul of medium containing 10% FBS was added to the lower chamber at 37°C for 48 h. Cells in the upper chamber were removed with cotton swabs, stained with crystal violet, photographed and counted.
Wound Healing Assay
A pipette tip was used to scratch a line on the cell layer and make marker. Washed two times with PBS and add 500ul of complete medium. Photographs were taken at 0h and 24h respectively after scratching and measured the width of the scratch.
Results
Analysis of DEGs
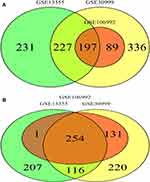
We utilized the Affy and Limma packages to analyze the three GEO data sets. The Venn diagram, consisting of 197 DEGs, from the three data sets, is illustrated in Figure 1A. The DEGs were further analyzed using the STRING database to identify potential links with other DEGs (Figure 1B). An interaction network diagram for these genes was additionally generated (Figure 2A). Figure 2B illustrates each DEG as a node and presents the degrees of each node. Based on this analysis, the degrees of the CXCL8 gene were found to be the largest, suggesting an essential role of CXCL8 in psoriasis. Hence, CXCL8 was selected for further analysis.
 |
Figure 2 (A) PPI network and the most significant module of DEGs. (B) Degree distribution of 197 DEGs. |
Analyzing Single Gene Difference
Next, we separated all samples into a high- or low-expressing group, based on their level of CXCL8 expression, and analyzed the differences between the groups (Figure 3). The GSE13355 data set had 62 high-expressing genes and 62 low-expressing genes. Likewise, the GSE30999 data set had 82 high-expressing genes and 82 low-expressing genes. Lastly, the GSE106992 data set had 96 high- and 96 low-expressing genes. Moreover, the heat map showed a marked difference in CXCL8 levels between the high- and low-expressing groups. Finally, based on the volcano plot, most genes were down-regulated in the GSE13355 and GSE30999 data sets, while the remaining genes were up-regulated in GSE106992.
CXCL8 Enrichment Analysis
The Venn diagram of 254 low- and high-expressing co-DEGs from the three data sets is shown in Figure 2B. In addition, we performed GO enrichment analysis and KEGG enrichment analysis on these genes (Figure 4). The GO analysis showed that these genes were markedly enriched in the “defense response to virus”, “type I interferon signaling pathway”, and “cell response to type I interferon” categories. Moreover, based on the KEGG analysis, these genes are markedly enriched in the “NOD-like receptor signaling pathway”, “interaction between cytokine and cytokine receptor”, and “IL−17 signaling pathway” categories (Table 1).
 |
Table 1 GO and KEGG Pathway Enrichment Analysis of DEGS in Psoriasis Samples |
 |
Figure 4 GO and KEGG pathway enrichment analysis of CXCL8 in psoriasis samples. (A) GO-BP (B) GO-MF (C) GO-CC (D) KEGG. |
GSEA and Analysis of Pan-Cancer Pathway
The biological pathways that were significantly altered in the CXCL8 high-expressing verses low-expressing group were determined using GSEA. These pathways were mainly related to the complement-like network (Figure 5A). To examine CXCL8 pathway enrichment in each cancer type, pan-cancer pathway analysis of CXCL8 was performed (Figure 5B) to identify the distribution pattern of CXCL8 gene in pan-cancer.
 |
Figure 5 (A) GSEA analysis of CXCL8. (B) Pan-cancer pathway of CXCL8. |
Immune Infiltration Analysis
We analyzed infiltrating immune cells between psoriatic and healthy skin specimen which are shown great differences in T cells regulatory, NK cells resting, monocytes and neutrophils. The analytical conclusions from data sets GSE13355, GSE30999, and GSE106992 are provided in Figures 6–8 respectively.
ROC Analysis
To better elucidate the relationship between CXCL8 expression and classification of diseased verses healthy tissues, we performed ROC analysis of the CXCL8 gene among the 3 data sets (Figure 9). The area under the ROC curves was 0.941 for GSE13355, as shown in Figure 9A. The area under the ROC curve was 0.935 for GSE30999, as shown in Figure 9B. The area under the ROC curve was 0.794 for GSE106992, as shown in Figure 9C. It can be seen that CXCL8 expression has a very good classification efficiency for both psoriatic and healthy skin specimen (AUC >0.7), indicating that CXCL8 can be used as a biomarker for psoriasis.
Constructing the ceRNA Network
Integrating starBase, miRDB, and miRWalk, 6 miRNAs, namely, hsa-miR-1294, hsa-miR-140-3p, hsa-miR-185-5p, hsa-miR-4306, hsa-miR-4644, and hsa-miR-493-5p, were predicted to interact with CXCL8 (Figure 10A). StarBase was further used to predict possible lncRNA interactions with any of the 6 six miRNAs (Figure 10C-H). Finally, using these data, a ceRNA network of CXCL8 gene was constructed (Figure 10B).
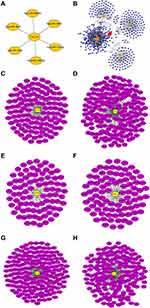 |
Figure 10 (A) Interactive network of CXCL8-miRNA. (B) CeRNA network of CXCL8-miRNA-LncRNA. (C–H) Six interactive networks of miRNA-LncRNA. |
HIC Analysis
The CXCL8 gene was further analyzed, using the HPA database, to obtain CXCL8 gene expression and distribution patterns throughout the human body. We discovered that CXCL8 was most highly expressed within the lymph node. We, thus, selected the lymph node for HIC analysis, and downloaded the HIC map of the CXCL8 gene in the lymph node (Figure 11).
 |
Figure 11 Immunohistochemical study on lymph node of CXCL8. (A-C) Images from three different patients. |
CXCL8 Influences the Inflammation Development of Psoriasis by Promoting Cell Proliferation, Migration, and Anti-Apoptosis
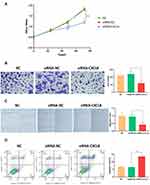
To evaluate the role of CXCL8 in terms of the proliferation and migration ability, we performed HaCat cell-line based assays. The siRNA-CXCL8 group significantly inhibited cell proliferation compared to the siRNA-NC group (48h, P<0.05; 72h, P<0.001, Figure 12A). Transwell assays showed that CXCL8 increased the ability of keratinocytes to invade remotely (P<0.05, Figure 12B). Similarly, the same trend was observed in the wound-healing assay, where the cell migration rate and distance were significantly attenuated when CXCL8 was inhibited (P<0.05, Figure 12C). Finally, analysis using flow cytometry showed that the siRNA-CXCL8 group had a significant pro-apoptotic effect (P<0.001, Figure 12D).
Discussion
Psoriasis is a highly prevalent chronic inflammatory skin disease. According to the latest epidemiological survey, the annual incidence rate of psoriasis in European countries is around 0.96 per thousand.38 Moreover, research revealed that the quality of life of patients with psoriasis is comparable to that observed in patients with different chronic internal diseases, such as arterial hypertension, diabetes mellitus, and cancer.39 Unfortunately, despite the availability of various treatments for psoriasis, this disease remains incurable. As a result, the development of highly effective and specific psoriasis biomarkers is imminent.
In this study, bioinformatics analysis of three separate microarray data sets with a total of 265 psoriasis patients and 213 healthy samples was used to identify genes specific for the pathogenesis of psoriasis. The STRING database was used for the analysis to verify the links between the DEGs, and 30 DEGs were selected as hub genes carrying degrees ≥ 20. CXCL8 exhibited the largest node degrees at 76, indicating its important role in the development and/or progression of psoriasis.
A number of studies have demonstrated CXCL8 to be an essential cytokine. It usually regulates tumor growth and invasion, and can be found in excess in tissues of patients with skin squamous cell carcinoma.40 Earlier reports also indicated that CXCL8 regulates the formation of Munro microabscess in psoriasis. Therefore, we evaluated CXCL8 expression in patients with psoriasis. Given that the role of CXCL8 in psoriasis has not been fully elucidated, we also explored the underlying function of CXCL8 in psoriasis.
In addition, using GO enrichment analysis, we demonstrated that in response to the virus, the type I interferon axis, and the corresponding cellular response, alterations in the markedly obvious modules occurred. In contrast, using KEGG, it was shown that the DEGs were mainly located on the NOD-like receptor axis, which is the interaction between cytokine and cytokine receptors and the IL−17 axis. Multiple studies have reported a strong relationship between the IL−17 axis and psoriasis. Il-17 signaling pathway plays a key role in the pathogenesis of psoriasis. Il-17a /F can activate keratinocytes to produce cytokines and interleukins, such as IL-8, which has a strong chemotaxis effect on neutrophils. Meanwhile, IL-17A/F can up-regulate chemokines expressed by keratinocytes (such as CXCL-1, CXCL-3, CXLC-5, and CXCL-6). This process is also linked to the action of neutrophils. In psoriasis, IL-17A also induces keratinocytes to express CCL-20, recruiting more Th17 cells and dendritic cells into the skin.41 In addition, IL-17A stimulates dendritic cells and fibroblasts to produce IL-6, further promoting T cells to differentiate into Th17.42 Il-17 promotes endothelial cell proliferation and angiogenesis by stimulating fibroblast production of vascular endothelial growth factor.In summary, these pieces of evidence suggest that IL-17A is closely related to the progression of psoriasis.
Using single-gene ROC analysis, we revealed that CXCL8 has great potential to serve as a biomarker for the diagnosis of psoriatic (AUC values>0.7). In addition, based on the CXCL8 predictions from starBase, miRDB, and miRWalk, we generated a CXCL8-ceRNA network, including hsa-miR-1294, hsa-miR-140-3p, hsa-miR-185-5p, hsa-miR-4306, hsa-miR-4644, and hsa-miR-493-5p. lncRNAs modulate immune and inflammatory pathways through multi-mechanistic regulation of gene expression.43 Among these mechanisms, the ceRNA axis has been extensively investigated. Based on the ceRNA theory, lncRNA activates target gene expression by sequestering miRNA that would otherwise suppress gene activation.44 Qiao et al,45 for instance, revealed a lncRNA-MSX2P1 axis that stimulates S100A71 and facilitates IL-22-induced keratinocytes production by inhibiting miR-6731-5p action. Similarly, Li et al46 reported that lncRNA H19 modulates the differentiation of keratinocytes by sequestering miR-130b-3p and augmenting desmoglein 1 expression. Consistent with these studies, our research also demonstrated a strong involvement of ceRNA in the development and progression of psoriasis through sequestering multiple potential miRNAs.
CXCL8 is most widely distributed in lymphoid tissues. Combined with the results of immune infiltration analysis, the differences in infiltrating immune cells between psoriasis patients and healthy skin specimens showed great differences in T cells regulatory, NK cells resting, monocytes, and neutrophils. Previous researchers have reported that the infiltration of T cells regulatory, NK cells resting, monocytes, and neutrophils plays an important role in the pathogenesis of psoriasis. Tominaga47 reported that CXCL8 can trigger the chemotaxis of neutrophils through CXCR2 and CXCR1.
Nevertheless, we delineated the role of CXCL8 in psoriasis by assessing its overall expression profile of CXCL8 in psoriasis and its potential mechanism of action. The RT-qPCR results of CXCL8 indicate that CXCL8 affects the inflammatory development of psoriasis by promoting cell proliferation, migration, and anti-apoptosis. Previous studies have shown that CXCL8 affects the occurrence and development of colorectal cancer, hepatocellular carcinoma, breast cancer, lung cancer, and thyroid tumor by regulating angiogenesis, tumor cell movement, immune cell infiltration, and tumor cell survival, as well as changing local anti-tumor immune responses.48 Chemokines are a class of small proteins involved in the transfer of leukocyte chemokines to inflammation sites, and CXCL8 is an important member of the CXC family. CXCL8 can promote the proliferation and invasion of colon cancer cells, and its mechanism may be closely related to the PI3K/Akt/ NF-KB signaling pathway. Although the RT-qPCR result is positive, the relationship between CXCL8 and psoriasis warrants further in-depth study.
Finally, some restrictions should be taken into consideration. Firstly, the stratification analysis and the interaction between factors have not been investigated in detail. In addition, due to its retrospective nature, we cannot rule out selectivity and recall bias. Therefore, we propose future prospective studies to improve statistical capabilities and achieve more reliable conclusions. Secondly, in addition to bioinformatics as a powerful tool for genome-wide research, insufficient understanding of the underlying mechanisms may limit our identification of effective psoriasis biomarkers. Therefore, CXCL8 must be extensively examined at the molecular, cellular, and organismal levels to confirm its significance in the pathogenesis of psoriasis.
Conclusions
In summary, this study analyzed 265 psoriatic skin lesion samples to identify DEGs responsible for psoriasis progression. We revealed 197 total DEGs associated with psoriasis and we recognized CXCL8 as a key biomarker for psoriasis. Cytological experimental validation has shown that CXCL8 plays a role in psoriatic inflammatory lesions by promoting cell proliferation, migration, and anti-apoptosis. Further investigations are warranted to explore all aspects of CXCL8 function in psoriasis.
Data Sharing Statement
All data generated or analysed during this study are included in this published article.
Ethics Approval
The study was approved by the Ethics Review Committee of Shanghai Skin Disease Hospital.
Acknowledgment
A preprint has previously been published (DOI: 10.21203/rs.3.rs-516404/v1).49
Funding
This work was sponsored by grants from National Natural Science Foundation of China (No. 81872522, 82073429), Innovation Program of Shanghai Municipal Education Commission (No.2019-01-07-00-07-E00046), Clinical Research Plan of SHDC (No. SHDC2020CR1014B, SHDC12018X06) and Program of Shanghai Academic Research Leader (No. 20XD1403300).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Larsen MH, Krogstad AL, Wahl AK. Alexithymia, illness perception and self-management competency in psoriasis. Acta Dermato Venereologica. 2017;97(8):934–940.
2. Gupta MA, Simpson FC, Gupta AK. Psoriasis and sleep disorders: a systematic review. Sleep Med Rev. 2016;29:63–75.
3. Ryan C, Kirby B. Psoriasis is a systemic disease with multiple cardiovascular and metabolic comorbidities. Dermatol Clin. 2015;33:41–55.
4. Nowowiejska J, Baran A, Flisiak I. Psoriasis and cardiometabolic disorders. Dermatol Rev. 2020;107:508–520.
5. Nowowiejska J, Baran A, Flisiak I. Aberrations in lipid expression and metabolism in psoriasis. Int J Mol Sci. 2021;22:6561.
6. Jiang S, Hinchliffe TE, Wu T. Biomarkers of an auto immune skin disease-psoriasis. Genom Proteom Bioinf. 2015;13:224–233.
7. Brodmerkel C, Li K, Garcet S, et al. Modulation of inflammatory gene transcripts in psoriasis vulgaris: differences between ustekinumab and etanercept. J Allergy Clin Immunol. 2019;143(5):1965–1969.
8. Ricceri F, Pescitelli L, Lazzeri L, et al. Clinical experience with infliximab biosimilar in psoriasis. Br J Dermatol. 2017;177(6):e347–e348.
9. Burmester GR, Gordon KB, Rosenbaum JT, et al. Long-Term Safety of Adalimumab in 29,967 Adult Patients From Global Clinical Trials Across Multiple Indications: an Updated Analysis. Adv Ther. 2020;37(1):364–380.
10. Carrascosa JM, Del-Alcazar E. New therapies versus first-generation biologic drugs in psoriasis: a review of adverse events and their management. Expert Rev Clin Immu. 2020;14(4):259–273.
11. Peranteau AJ, Turkeltaub AE, Tong Y, et al. A Review of Ixekizumab, an Anti-Interleukin-17A Monoclonal Antibody, for Moderate-to-Severe Plaque Psoriasis. Skin Therapy Lett. 2016;21(6):1–6.
12. Feldman SR, Gomez B, Meng X, et al. Secukinumab rapidly im-proves EQ-5D health status in patients with psoriasis: pooled analysis from four Phase 3 trials. J Dermatology Treat. 2020;1:1–7.
13. Roy I, Evans DB, Dwinell MB. Chemokines and chemokine receptors: update on utility and challenges for the clinician. Surgery. 2014;155(6):961–973.
14. Palomino DC, Marti LC. Chemokines and immunity. Einstein. 2015;13(3):469–473.
15. Timoteo RP, Silva MV, Miguel CB, et al. Th1 /Th17-related cytokines and chemokines and their implications in the pathogenesis of pemphigus vulgaris. Mediators Inflamm. 2017;22(2):715–1285.
16. Helen H, Bikash D, Nouri N. Role of the CXCL8-CXCR1 /2 axis in cancer and inflammatory diseases. Theranostics. 2017;7(6):1543–1588.
17. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995.
18. Liberzon A, Birger C, Thorvaldsdóttir H, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425.
19. Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38(6):675–678.
20. Gautier L, Cope L, Bolstad BM, et al. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315.
21. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47.
22. Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011;26(12):35.
23. Russell CB, Rand H, Bigler J, et al. Gene expression profiles normalized in psoriatic skin by treatment with brodalumab, a human anti‑IL‑ 17 receptor monoclonal antibody. J Immunol. 2014;192:
24. Bigler J, Rand HA, Kerkof K, et al. Cross-study homogeneity of psoriasis gene expression in skin across a large expression range. PLoS One. 2013;8:e52242.
25. Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287.
26. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550.
27. Von Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31(1):258–261.
28. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504.
29. Ito K, Murphy D. Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacometrics Syst Pharmacol. 2013;2:e79.
30. Chen B, Khodadoust MS, Liu CL, et al. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259.
31. Akobeng AK. Understanding diagnostic tests 3: receiver operating characteristic curves. Acta Paediatrica. 2007;96(5):644–647.
32. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12(1):77.
33. Jeggari A, Marks DS, Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28(15):2062–2063.
34. Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7.
35. Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48:D127–D131.
36. Dweep H. Gretz N, and Sticht C, miRWalk database for miRNA-target interactions. Methods Mol Biol. 2014;1182:289–305.
37. Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue Based Map Human Proteome Sci. 2015;347(6220):1260419.
38. Valent F, Tullio A, Errichetti E, et al. The epidemiology of psoriasis in an Italian area: population-based analysis of administrative data. G Ital Dermatol Venereol. 2018;29(6):867.
39. Martı´nez JM, Nogueras P, Mun˜oz-Negro JE, et al. Quality of life, anxiety and depressive symptoms in patients with psoriasis: a case-control study. J Psychosom Res. 2019;124:109780.
40. ARTUC M, GUHL S, BABINA M, et al. Mast cell-derived TNF-alpha and histamine modify IL-6 and IL-8 expression and release from cutaneous tumor cells. Exp Dermatol. 2011;20(12):1020–1022.
41. Conti HR, Gaffen SL. IL-17-mediated immunity to the opportunistic fungal pathogen candida albicans. J Immunol. 2015;195(3):780–788.
42. Carrier Y, Ma HL, Ramon HE, et al. Inter-regulation of Th17 cytokines and the IL-36 cytokines in vitro and in vivo: implications in psoriasis pathogenesis. J Invest Dermatol. 2011;131(12):2428–2437.
43. Wu GC, Pan HF, Leng RX, et al. Emerging role of long noncoding RNAs in autoimmune diseases. Auto Immune Rev. 2015;14:798–805.
44. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352.
45. Qiao M, Li R, Zhao X, et al. Up-regulated lncRNA-MSX2P1 promotes the growth of IL-22-stimulated keratinocytes by inhibiting miR-6731-5p and activating S100A7. Exp Cell Res. 2018;363:243–254.
46. Li CX, Li HG, Huang LT, et al. H19 lncRNA regulates keratinocyte differentiation by targeting miR-130b-3p. Cell Death Dis. 2017;8:e3174.
47. Tominaga M, Okamoto M, Kawayama T, et al. Overexpression of IL-38 protein in anti cancer drug-induced lung injury and acute exacerbation of idiopathic pulmonary fibrosis. Respir Investig. 2017;55(5):293–299.
48. Liu Q, Li A, Tian Y, et al. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 2016;31:61–71.
49. Yang Y, Xie S, Lu J, Tang S. Screening and Identification of Potential Key Biomarkers in Psoriasis: evidence From Bioinformatic Analysis. Res Square. 2021;2:9867.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.