Back to Journals » Infection and Drug Resistance » Volume 15
Differentially Expressed Genes of Pseudomonas aeruginosa Isolates from Eyes with Keratitis and Healthy Conjunctival Sacs
Authors Ma X, Liu Q, Song F, Huang Y
Received 19 May 2022
Accepted for publication 6 August 2022
Published 12 August 2022 Volume 2022:15 Pages 4495—4506
DOI https://doi.org/10.2147/IDR.S374335
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Xiubin Ma,1– 3 Qing Liu,1– 3 Fangying Song,1– 3 Yusen Huang1– 3
1Department of Ophthalmology, Eye Institute of Shandong First Medical University, Qingdao Eye Hospital of Shandong First Medical University, Qingdao, People’s Republic of China; 2State Key Laboratory Cultivation Base, Shandong Provincial Key Laboratory of Ophthalmology, Shandong Eye Institute, Qingdao, People’s Republic of China; 3Department of Ophthalmology, School of Ophthalmology, Shandong First Medical University, Qingdao, People’s Republic of China
Correspondence: Yusen Huang, Department of Ophthalmology, Qingdao Eye Hospital of Shandong First Medical University, Qingdao, 266071, People’s Republic of China, Tel +86-532-85876380, Email [email protected]
Background: Pseudomonas aeruginosa (P. aeruginosa) is the second-most common commensal bacterium in healthy conjunctival sacs. When the corneal epithelial barrier is damaged, P. aeruginosa in a healthy conjunctival sac can cause infectious keratitis, which can result in the loss of vision. This study was designed to investigate the differentially expressed genes (DEGs) of P. aeruginosa isolates from eyes with keratitis and from healthy conjunctival sacs to predict their functions and pathways through Illumina high-throughput RNA sequencing (RNA-seq).
Methods: P. aeruginosa isolates from keratitis and healthy conjunctival sacs were obtained. The transcriptome profile of P. aeruginosa was characterized by a high throughput RNA-seq strategy using the Illumina HiSeq 2500 platform. The DEGs were analyzed with DESeq and validated through quantitative real-time polymerase chain reaction (PCR) and with experimental mice. GO enrichment and the KEGG pathway were also analyzed.
Results: In genome-wide transcriptional analysis, 557 genes (332 upregulated and 225 downregulated) were found to be differentially expressed (fold change ≥ 2, p ≤ 0.05) in the strains from keratitis. GO enrichment analysis suggested that DEGs tended to be associated with cellular and metabolic processes. KEGG pathway analysis revealed the DEGs were typically associated with the pathways of the bacterial secretion system and pyoverdine metabolism. Eleven DEGs were validated using quantitative reverse-transcription PCR and verified with experimental mice. The results were consistent with those obtained in RNA-seq.
Conclusion: The DEGs related to pilin, T2SS, T3SS, and pyoverdine metabolisms were significantly altered in the strains from keratitis. The findings may be helpful for further investigations on genes or pathways related to the pathogenesis of and therapeutic targets for P. aeruginosa keratitis.
Keywords: P. aeruginosa, genotype, keratitis, conjunctival sac, microflora
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is the second-most common commensal bacterium in healthy conjunctival sacs1 and can easily contaminate the cornea. It produces a series of cell-associated and extracellular virulence factors, such as pili, flagella, elastase, exotoxin A, a type III secretion system (T3SS), and others, and it can invade or kill corneal cells and induce corneal destruction.2,3 When the corneal epithelial barrier is damaged by injury or contact lens wear, P. aeruginosa can cause infectious keratitis.4,5 P. aeruginosa keratitis is an invasive corneal infection and a leading cause of blindness worldwide.6–8 The pathogenesis of P. aeruginosa keratitis involves bacterial virulence and induces excessive inflammatory response in the host. Its resistance to antibiotics is increasing via intrinsic and acquired mechanisms,9–12 and the treatment of P. aeruginosa keratitis remains a thorny problem in clinical practice. As the severity of keratitis and its sensitivity to medication depend on the virulence factors of the pathogen, additional insights into P. aeruginosa are required. Therefore, it is important to investigate similarities and differences in characteristics among P. aeruginosa isolates from eyes with keratitis and those from healthy conjunctival sacs.
While genome analysis of P. aeruginosa strains (such as PAO1, PA19660, and PaeAG1) have been conducted,13–17 they have generally been limited to differences in phenotypic and genetic characterization. The advent of next-generation sequencing has made whole-genome, transcriptome, and even epigenomic sequencing of organisms possible. A comprehensive understanding of the transcriptome might contribute to the discovery of the functional elements of the genome and to hypotheses regarding potential mechanisms in physiological and pathological conditions.18 High-throughput RNA sequencing (RNA-seq) has become the standard technique to analyze transcriptomes and an alternative to transcriptomic technologies such as microarrays because of its advantages in covering a large dynamic range, possessing a high level of reproducibility, and requiring fewer RNA samples.18 This tool has identified differences in gene expression between biological samples such as Escherichia coli, Salmonella typhi, and Helicobacter pylori.19–21 For example, Molina-Mora et al identified transcriptomic determinants of the response of ST-111 P. aeruginosa AG1 to ciprofloxacin by employing RNA-seq.22 We previously used the technique to identify the gene characteristics of Staphylococcus aureus isolated from healthy conjunctival sacs and eyes with postoperative endophthalmitis.23 In the current study, we aimed to investigate the differentially expressed genes (DEGs) of P. aeruginosa isolates from keratitis and healthy conjunctival sacs by using Illumina high-throughput RNA-seq technology. We also performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of the DEGs and associated pathway genes involved in the pathogenesis of, and thus potential therapeutic targets against, P. aeruginosa keratitis.
Materials and Methods
Bacterial Isolates
Four P. aeruginosa isolates from P. aeruginosa keratitis and three P. aeruginosa isolates from healthy conjunctival sacs were obtained from the Clinical Laboratory of the Qingdao Eye Hospital of Shandong First Medical University (Qingdao, China). Four strains of P. aeruginosa were isolated from four infected corneas with P. aeruginosa keratitis (patient information provided in Table 1) by cornea scraping and cultured in the Laboratory. Three strains of commensal P. aeruginosa were isolated from the conjunctival sacs of three individuals with healthy eyes (Table 1) who had not used local mydriatic agents or antibiotic drops for 1 month prior to swab collection of the specimens. No conjunctival, facial, or systemic infections were found. All the samples were inoculated on blood agar plates (Auto Biotechnology, Zhengzhou, China) in triplicate and incubated for 24 h. Incubated specimens with a single-colony morphology were identified by an automatic microbiological identification and susceptibility analysis system (Beckman Coulter WalkAway-96 plus, CA, USA) using colorimetry and fluorescence to identify both gram-negative and gram-positive organisms through a variety of biochemical reactions. One single-colony morphology was selected to be cultured in 10 mL of Luria-Bertani (LB) broth at 37°C and grown to a turbidity of 0.5 at 600 nm, with shaking at 160 × rpm. The bacterial cultures were centrifuged, washed, and collected for follow-up analysis. The bacterial preparation procedures were performed in accordance with a previously reported protocol.23
 |
Table 1 Characteristics of the Four Patients with Keratitis and Three Healthy Individuals |
The study was approved by the Ethics Committee of Qingdao Eye Hospital of Shandong First Medical University (No. 2020-G-12). In accordance with the tenets of the Declaration of Helsinki, all enrolled patients provided written informed consent prior to undergoing the procedure.
RNA-Seq Library Construction and Sequencing
The bacterial collection was added with phenol water (Sinopharm Chemical Reagent Co., Shanghai, China) and shaken violently to be mixed. The centrifuge tube was incubated at 65°C with an oscillating metal bath at maximum speed for 30–60 minutes, cooled on ice for 5 minutes, then centrifuged at 4°C for 10 minutes. The upper water phage was absorbed and added with 1/2 volume of TRK-1002 lysate (LC-Bio Technology CO., Ltd, Hangzhou, China). The concentration and purity of RNA were measured using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). An RNA integrity number (RIN) >7.0 can be used to construct a cDNA library, and the cDNA library was prepared and sequenced by Lianchuan Bio (Hangzhou, China). Ribosomal RNA were removed from the total RNA by using a Ribo-Zero Magnetic Kit (Epicentre, Madison, WI, USA), and then the library was generated using an Illumina Truseq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s recommendation. After digested into 200 nt fragments, double-stranded cDNAs were synthesized using uracil-N-glycosylase (UNG) to a cDNA cluster. The cDNAs were used to create the RNA-seq library by following the standard Illumina protocol (Lianchuan Bio, Hangzhou, China).
Analysis of RNA-Seq Data
The raw paired-end reads were trimmed and quality controlled by Trimmomatic with the default parameters (http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic). Then, clean reads were separately aligned to the reference genome (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE209539) in orientation mode using Rockhopper (http://cs.wellesley.edu/~btjaden/Rockhopper/) software. Rockhopper is a comprehensive, user-friendly system for computational analysis of bacterial RNA-seq data.24 As input, Rockhopper takes RNA sequencing reads generated by high-throughput sequencing technology. This software was used to calculate gene expression levels with the default parameters. To identify DEGs between the two samples, the expression level for each transcript was calculated using the fragments per kilobase of read per million mapped reads (RPKM) method.
DEG analysis of the P. aeruginosa isolates was performed by LC-Bio Technology CO., Ltd (Hangzhou, China) using edgeR (https://bioconductor.org/packages/release/bioc/html/edgeR.html). The false discovery rate was controlled by adjusting the p values using the Benjamini-Hochberg approach. Genes with an adjusted p value of <0.05 were considered DEGs. To obtain significant functions of the DEGs, a GO enrichment analysis was performed using the GO seq R package. Pathway analysis was performed using the KEGG pathway database (http://www.genome.jp/kegg).
Quantitative Reverse-Transcription Polymerase Chain Reaction
Eleven genes from the significantly enriched KEGG pathway were chosen for quantitative reverse-transcription polymerase chain reaction (qRT-PCR), which showed the same expression trend in all strains of the same group (|log2 fold change| > 2, adjusted p < 0.05) (Table 2). Mouse corneas (see below) infected with different P. aeruginosa isolates were excised, minced, and homogenized in 100 µL of PBS with a TissueLyser (TissueLyser II, Hilden, Germany). The total RNA samples were reversed to generate cDNA using a PrimeScript RT Reagent Kit. Realtime PCR was conducted using TB Green Premix EX Taq II (TaKaRa, Beijing, China) and Rotor-Gene Q systems (Qiagen, Hilden, North Rhine-Westphalia, Germany). The cycling conditions were 30s at 95°C followed by two-step cycles (5s at 95°C and 1 min at 60°C). The quantified data were analyzed using the ΔΔ threshold cycle method with 16s rRNA as an internal control for P. aeruginosa. The primers are shown in Supplementary Table 1.
 |
Table 2 Genes Validated by Quantitative RT-PCR |
Animals
Female C57BL/6 mice (age, 8 weeks; weight, 20–25 g; amount, 90) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd., and housed in the animal center of Qingdao Eye Hospital of Shandong First Medical University. All procedures were performed in accordance with the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Ophthalmic and Vision Research. The study was conducted with the approval of the Ethics Committee of Qingdao Eye Hospital of Shandong First Medical University (No. 2020-G-12).
Infection Procedure and Clinical Examination
Each experiment was repeated thrice, with five eyes used in each group. The mice were anesthetized with ether, and the cornea of the left eye was scratched to create three 1 mm incisions by using a sterile 25-gauge needle and inoculated with bacterial suspension containing 1×106 colony-forming units (CFU) of the P. aeruginosa isolates from the healthy conjunctival sac group. The procedures were performed in accordance with a previously reported protocol.4 The corneas were examined with a slit lamp at 24, 48, and 72 h after inoculation. The severity of the corneal disease was graded using a well-established scale.25 At 24, 48, and 72 h after inoculation, and the infected corneas were cut along the limbus under a microscope immediately after the mice were euthanized.
Determination of Bacterial Load
Each cornea was homogenized with a tissue homogenizer (TissueLyser II, Hilden, Germany) in sterile saline. Aliquots (100 mL) of serial dilutions were plated onto LB agar plates in triplicate and cultured at 37°C for 24 h. The bacterial colonies were counted and recorded as CFU per cornea.
Statistical Analysis
A statistical analysis qRT-PCR, clinical score, and bacterial load data was performed using SPSS 20.0 software (SPSS Software, Chicago, IL, USA). An unpaired, two-tailed Student’s t-test and the Mann–Whitney U-test were performed. Data obtained from at least three experiments are expressed as mean ± standard deviation (SD). Differences were considered statistically significant at p < 0.05 (*p < 0.05, **p < 0.01, and ***p < 0.001).
Results
DEGs Analysis
To investigate changes in the gene expressions of P. aeruginosa isolates from keratitis and healthy conjunctival sac groups, the DEGs were analyzed. Gene expression was considered differential, with a corrected p value of <0.05. A total of 557 DEGs were identified, of which 332 genes (59.6%) were upregulated and 225 genes (40.4%) downregulated in the keratitis group. Scatter and volcano plots were used to visualize the characteristics of the DEGs (Figure 1A and B).
GO Functional Analysis of DEGs
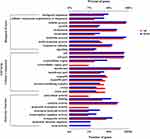
GO enrichment analysis was used to ascribe the three main categories of gene functions, namely biological process, molecular function, and cellular component. It was performed to analyze the functions of all the DEGs in the keratitis and healthy conjunctival sac groups by using Fisher’s exact test with p ≤ 0.05 as the threshold. Among 53 GO terms (Supplementary Table 2), 39 GO terms were classified as biological processes, 10 GO terms as molecular functions, and 4 GO terms as cellular components. As shown in Figure 2 and Supplementary Table 2, for biological processes, the top two GO terms were “peptide transport (GO:0015833)” and “amide transport (GO:0042886)”. “Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor (GO:0016708)” and “5-carboxymethyl-2-hydroxymuconate delta-isomerase activity (GO:0008704)” were the top two GO terms for molecular functions, and “extracellular region (GO:0005576)”, “cell projection (GO:0042995)”, and “type II protein secretion system complex (GO:0015627)” were the top three GO terms for cellular components. Moreover, the genes (“protein secretion by the type III secretion system, GO:0030254” and “type III protein secretion system complex, GO:0030257”) involved with the T3SS were the more significantly changed DEGs, indicating that the T3SS may play an important role in the pathogenesis of P. aeruginosa-induced keratitis.
KEGG Pathway Analysis of DEGs
To investigate the pathways involved in the P. aeruginosa isolates, a KEGG pathway analysis was performed for the keratitis group, which was compared with the healthy conjunctival sac group. A total of 59 pathways are shown in Figure 3. The bacterial secretion system was the most significantly enriched upregulated pathway, indicating that compared with P. aeruginosa in healthy conjunctival sacs, the activity of P. aeruginosa secretion function in keratitis was significantly enhanced. The most significantly enriched downregulated pathways were the pyoverdine metabolic and biosynthetic processes, indicating that iron metabolism in P. aeruginosa during keratitis was relatively weakened.
Validation of DEGs with qRT Polymerase Chain Reaction
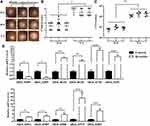
To validate the RNA-seq results, qRT-PCR was performed using the same extracted total RNA as in the RNA-seq analysis for each of the seven samples. A total of 11 genes (|log2 fold change| > 2; adjusted p < 0.05) from the significantly enriched KEGG pathway were validated. The 11 genes exhibited the same expression trend in all strains of the same group (Table 2). Of the 11 validated genes, 9 were upregulated and involved with the bacterial secretion system and pilin of the significantly enriched KEGG pathway, and 2 genes were downregulated and involved with the pyoverdine metabolic process of the significantly enriched KEGG pathway in the keratitis group, as indicated by the RNA-seq results. Of the nine upregulated genes, two were involved with the T2SS and two with pilin and thus involved with motility of Pseudomonas, whereas the other five were involved with the T3SS, which is an important pathogenic virulence factor of P. aeruginosa. In our validated results, the expression levels of the nine genes involved with the bacterial secretion system and pilin were significantly upregulated, and those of the two genes involved with the pyoverdine metabolic process were significantly downregulated in the keratitis group compared with the healthy conjunctival sac group. The results were consistent with the RNA-seq results, indicating the reliability of the latter (*p < 0.05, **p < 0.01, and ***p < 0.001; Figure 4).
Changes of the DEGs of P. aeruginosa Isolates from Eye with Healthy Conjunctival Sac to Keratitis
To further verify the changes of DEGs of P. aeruginosa isolates from eyes with a healthy conjunctival sac to keratitis, isolates from healthy sacs were incubated and inoculated onto the surface of murine scarified corneas with 1.0×106 CFU, and infectious keratitis was induced. The expression of DEGs of P. aeruginosa in infectious keratitis and that of P. aeruginosa before inoculation was detected. Representative photographs (Figure 5A) showed that the corneas treated with P. aeruginosa isolates from the healthy conjunctival sac showed slight corneal opacification at 24 h and were nearly perforated at 72 h after bacterial inoculation. The gradual increase in clinical scores also indicated that the destruction of infected corneas was aggravated gradually (Figure 5B). The bacterial load of infected corneas at 24 and 72 h after inoculation was also measured (Figure 5C). The expression of the 11 DEGs chosen from the significantly enriched KEGG pathway (Figure 5D) from murine infectious keratitis at 48 h after inoculation and healthy conjunctival sacs was detected. The results showed that two DEGs involved with the pyoverdine metabolism were downregulated, and nine DEGs involved with the bacterial secretion system and pilin were upregulated, which was consistent with the RNA-seq results. The results also indicated changes in the DEGs of P. aeruginosa isolates from eyes with a healthy conjunctival sac to keratitis.
Discussion
P. aeruginosa-colonized healthy conjunctival sacs can contaminate the ocular surface and lead to keratitis when the corneal epithelium is damaged. To understand the pathogenesis of keratitis, the relationship between P. aeruginosa isolated from healthy conjunctival sacs and that isolated from eyes with keratitis must be investigated. In the present study, we found 557 DEGs between the P. aeruginosa isolates from keratitis and healthy conjunctival sac groups. Our results indicated dramatic changes in the bacterial secretion system and iron metabolic system-related genes. To the best of our knowledge, this is the first study using RNA-seq to investigate the relationship between commensal and keratitis-causing strains.
The GO functional annotation of the transcripts in the current study revealed that both upregulated and downregulated genes were related to the metabolic process. The results of enrichment analysis indicated that the terms of peptide transport and amide transport were significantly enriched in the biological process. By analyzing these involved genes, we found that most of the DEGs presented an upregulated trend. Specifically, the GO terms “protein secretion by the type III secretion system” and “type III protein secretion system complex” were associated with the T3SS and were significantly upregulated in the keratitis group. This indicated that the T3SS may play an important role in the pathogenesis of P. aeruginosa-induced keratitis and may be speculated as an attractive antibiotic target for treatment.
The results of KEGG pathway enrichment analysis indicated that pathways of the bacterial secretion system, pyoverdine metabolic, and biosynthetic processes enriched more DEGs. Gram-negative bacteria employ secretion systems to translocate proteinaceous effectors from the cytoplasm to the extracellular milieu, thereby interacting with the surrounding environment or microniche.26 In the present study, more DEGs were enriched in the T3SS, indicating that the T3SS most significantly changed the secretion status of P. aeruginosa in the keratitis group and was associated with the destruction to the cornea caused by P. aeruginosa. The pathways of pyoverdine metabolic and biosynthetic processes are involved in iron ion metabolism. Iron is an essential micronutrient for the growth and proliferation of all organisms, including pathogenic bacteria, and plays a pivotal role in colonization and subsequent pathogenesis.27 Iron limitation is a vital host defense mechanism against bacteria.
Various P. aeruginosa virulence factors are involved in the pathogenic process of corneal infection. Most play needless roles, but some are required for full virulence.28,29 The T2SS consists of an assembly of 12 to 15 Gsp proteins responsible for transporting a variety of virulence factors across the outer membrane in several pathogenic bacteria. It is a key virulence factor in P. aeruginosa. Many diverse effectors and toxins depend on the T2SS for secretion. Such substrates are involved in adhesion, biofilm formation, nutrient acquisition, colonization, and invasion.30 Pilin is a virulent protein on the surface of P. aeruginosa. The twitching motility produced by the extension and retraction of type IV pili assists bacterial adherence onto the corneal epithelium and enables bacterial infiltration into the corneal stroma, which is required for the pathogenic process of gram-negative bacteria.31,32 This mechanism also helps bacteria effectively escape from the surface when needed.33 Twitching motility mutants show a lack of virulence in vivo.34 A mutant of P. aeruginosa was shown to have a twitching motility defect and to exhibit decreased corneal colonization in mice,30 which suggests that type IV pili play an important role in corneal infection. A previous report demonstrated that 90% of 63 keratitis isolates showed better twitching motility than the PA14 strain.35 In gram-negative bacteria, type IV pilus assembly and T2SS polymerize inner membrane proteins called major pilins and pseudopilins, respectively, into thin filaments. Four minor pilins are required in both systems for efficient fiber assembly.30 In our results (Figures 4B and 5), compared with P. aeruginosa isolated from healthy conjunctival sacs, that isolated from eyes with keratitis was also found to carry more T2SS and type IV pilus genes, to be more virulent, and to induce more severe corneal infections.
The T3SS is a well-recognized major virulence determinant in P. aeruginosa. It conveys toxins to host cells and plays an important role in inducing host infection. It includes a pore-forming protein, needle-like devices, and effector proteins. The four effector toxins of T3SS that are involved in virulence are ExoS, ExoU, ExoT, and ExoY.36 ExoS and ExoT encode for bifunctional enzymes that comprise a GTPase activating domain and an ADP ribosyltransferase domain. ExoU encodes for a cytotoxin phospholipase A2, and ExoY encodes for adenylate cyclase. P. aeruginosa employs the T3SS for immune evasion and persistence determinants, such as Psl exopolysaccharide, to form tenacious biofilms.37,38 ExoS and ExoT are closely related and have been shown to block reactive oxygen species (ROS) production, induce neutrophil apoptosis, and inhibit neutrophil phagocytosis.39,40 The presence of ExoU is associated with increased cytotoxicity. ExoU+ isolates were previously reported to mediate pathogenicity in an experimental model of keratitis and to induce cell lysis in macrophages and epithelial cells. The T3SS has been associated with increased mortality and persistence in patients with acute lung infections.41 In keratitis, the adenosine diphosphate ribosyl transferase activities of Exo S and T mediate the subversion of the host immune response to promote bacterial survival and the development of corneal disease.42 In our study, we also found that the gene expression of T3SS (shown in Figure 4C) was significantly more upregulated in the keratitis group than in the healthy conjunctival sac group, confirming previous reports.42 The results indicated that it could be associated with the destruction caused by P. aeruginosa to the cornea.
Iron is indispensable in the in vivo growth of all human pathogens. The growth of P. aeruginosa is also regulated by iron and the quorum sensing system. Iron restriction in host fluids for proteins such as lactoferrin and transferrin is a vital defense mechanism against bacteria.43 Pyoverdines play an essential role in the iron metabolic process and is a critical virulence determinant of P. aeruginosa during acute infections. One pyoverdine uptakes iron and releases PvdS to regulate several virulence factors and the production machinery for pyoverdine itself.44 Another pyoverdine removes iron and causes mitochondrial damage.45 While Wiens et al46 observed that pyoverdine production contributed to P. aeruginosa biofilm formation, another study found no evidence that pyoverdine production had an effect on biofilm formation in various clinical and environmental isolates.47 During infection, pyoverdines contribute to the growth and virulence of P. aeruginosa by providing iron, a process hosts assiduously try to prevent. The expression of the acyl-homoserine lactone acylase PvdQ and that of pyoverdine non-ribosomal peptide synthase/polyketide synthase PvdL genes in our study were downregulated in the keratitis group (Figure 4A), perhaps due to host regulation.
This study has several limitations. First, we collected only four P. aeruginosa isolates from eyes with keratitis because the positivity rate in bacterial culture was not high. Second, gene regulation was studied in laboratory growth media, which may exhibit substantially different gene regulation than that in bacteria in the host environment. Third, several of the genes we identified as possibly contributing to the pathogenesis of keratitis may need to be validated in future studies. Fourth, P. aeruginosa is not a clonal bacterium, and there are variations in genome size within the group. Therefore, the use of a reference genome is not always the best option for Pseudomonas genome analysis. It is possible that a de novo genome assembly can offer new DEGs that correspond to exclusive genes in the isolate (that are relevant for biological observation) but are absent in the reference genome.22,48 Therefore, paying closer attention to and analyzing the genomes of clinical isolates remains necessary.
In conclusion, DEGs related to the pilin, T2SS, T3SS, and pyoverdine metabolisms significantly changed in the pathogenesis of P. aeruginosa of healthy conjunctival sacs to induced keratitis. This indicates that the virulence factors corresponding to these DEGs may play key roles in inducing keratitis by commensal P. aeruginosa in a healthy conjunctival sac. The development of novel measures against these virulence factors may play a better role in the prevention and treatment of P. aeruginosa keratitis in clinical practice.
Data Sharing Statement
All data relevant to the study are included in the article. The datasets used and analyzed during the current study are available from the corresponding authors at any time upon reasonable request.
Ethics Approval and Informed Consent Statement
The study was obtained from the approval of Ethics Committee of the Qingdao Eye Hospital of Shandong First Medical University and conducted followed the ethical standards presented in the 1964 Declaration of Helsinki and its later amendments. All the enrolled patients provided written informed consent prior to undergoing the procedure.
Acknowledgments
We thank Huabo Chen for kindly providing the bacterial isolates and thank Qi Xia for the technical support and guidance.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the Shandong Provincial Medicine and Health Science and Technology Development Program (202007020620) and the National Natural Science Foundation of China (82171027 and 81970788).
Disclosure
All authors declared no conflicts of interest in relation to this work.
References
1. Huang Y, Yang B, Li W. Defining the normal core microbiome of conjunctival microbial communities. Clin Microbiol Infect. 2016;22:643, e7–e12. doi:10.1016/j.cmi.2016.04.008
2. Suzuki T, Okamoto S, Oka N, Hayashi N, Gotoh N, Shiraishi A. Role of pvdE pyoverdine synthesis in Pseudomonas aeruginosa keratitis. Cornea. 2018;37:S99–S105. doi:10.1097/ICO.0000000000001728
3. Oka N, Suzuki T, Ishikawa E, et al. Relationship of virulence factors and clinical features in keratitis caused by Pseudomonas aeruginosa. Invest Ophthalmol Vis Sci. 2015;56:6892–6898. doi:10.1167/iovs.15-17556
4. Ma X, Wang Q, Song F, et al. Corneal epithelial injury-induced norepinephrine promotes Pseudomonas aeruginosa keratitis. Exp Eye Res. 2020;195:108048. doi:10.1016/j.exer.2020.108048
5. Li J, Ma X, Zhao L, Li Y, Zhou Q, Du X. Extended contact lens wear promotes corneal norepinephrine secretion and Pseudomonas aeruginosa infection in mice. Invest Ophthalmol Vis Sci. 2020;61:17. doi:10.1167/iovs.61.4.17
6. Lakhundi S, Siddiqui R, Khan NA. Pathogenesis of microbial keratitis. Microb Pathog. 2017;104:97–109. doi:10.1016/j.micpath.2016.12.013
7. Carnt N, Samarawickrama C, White A, Stapleton F. The diagnosis and management of contact lens-related microbial keratitis. Clin Exp Optom. 2017;100:482–493. doi:10.1111/cxo.12581
8. Sharma P, Elofsson M, Roy S. Attenuation of Pseudomonas aeruginosa infection by INP0341, a salicylidene acylhydrazide, in a murine model of keratitis. Virulence. 2020;11:795–804. doi:10.1080/21505594.2020.1776979
9. Austin A, Lietman T, Rose-Nussbaumer J. Update on the management of infectious keratitis. Ophthalmology. 2017;124:1678–1689. doi:10.1016/j.ophtha.2017.05.012
10. Cunrath O, Meinel DM, Maturana P, et al. Quantitative contribution of efflux to multi-drug resistance of clinical Escherichia coli and Pseudomonas aeruginosa strains. EBioMedicine. 2019;41:479–487. doi:10.1016/j.ebiom.2019.02.061
11. Subedi D, Vijay AK, Willcox M. Overview of mechanisms of antibiotic resistance in Pseudomonas aeruginosa: an ocular perspective. Clin Exp Optomet. 2018;101:162–171. doi:10.1111/cxo.12621
12. Vazirani J, Wurity S, Ali MH. Multidrug-resistant Pseudomonas aeruginosa keratitis: risk factors, clinical characteristics, and outcomes. Ophthalmology. 2015;122:2110–2114. doi:10.1016/j.ophtha.2015.06.007
13. Yamaguchi S, Suzuki T, Kobayashi T, et al. Genotypic analysis of Pseudomonas aeruginosa isolated from ocular infection. J Infect Chemother. 2014;20:407–411. doi:10.1016/j.jiac.2014.02.007
14. Lakshmi PJ, Prajna L, Mohankumar V. Genotypic and phenotypic characterization of Pseudomonas aeruginosa isolates from post-cataract endophthalmitis patients. Microb Pathog. 2015;78:67–73. doi:10.1016/j.micpath.2014.11.014
15. Doustdar F, Karimi F, Abedinyfar Z, Amoli FA, Goudarzi H. Genetic features of Pseudomonas aeruginosa isolates associated with eye infections referred to Farabi Hospital, Tehran, Iran. Int Ophthalmol. 2019;39:1581–1587. doi:10.1007/s10792-018-0980-5
16. Molina Mora JA, Montero-Manso P, García-Batán R, Campos-Sánchez R, Vilar-Fernández J, García F. A first perturbome of Pseudomonas aeruginosa: identification of core genes related to multiple perturbations by a machine learning approach. Biosystems. 2021;205:104411. doi:10.1016/j.biosystems.2021.104411
17. Molina-Mora JA, Chinchilla-Montero D, García-Batán R, García F. Genomic context of the two integrons of ST-111 Pseudomonas aeruginosa AG1: a VIM-2-carrying old-acquaintance and a novel IMP-18-carrying integron. Infect Genet Evol. 2021;89:104740. doi:10.1016/j.meegid.2021.104740
18. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi:10.1038/nrg2484
19. Li S, Dong X, Su Z. Directional RNA-seq reveals highly complex condition-dependent transcriptomes in E. coli K12 through accurate full-length transcripts assembling. BMC Genom. 2013;14:520. doi:10.1186/1471-2164-14-520
20. Perkins TT, Kingsley RA, Fookes MC, et al. A strand-specifc RNA-Seq analysis of the transcriptome of the typhoid bacillus Salmonella typhi. PLoS Genet. 2009;5:e1000569. doi:10.1371/journal.pgen.1000569
21. Sharma CM, Hoffmann S, Darfeuille F, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi:10.1038/nature08756
22. Molina-Mora JA, Chinchilla-Montero D, Chavarría-Azofeifa M, et al. Transcriptomic determinants of the response of ST-111 Pseudomonas aeruginosa AG1 to ciprofloxacin identified by a top-down systems biology approach. Sci Rep. 2020;10:13717. doi:10.1038/s41598-020-70581-2
23. Liu Q, Chen N, Chen H, Huang Y. RNA-Seq analysis of differentially expressed genes of Staphylococcus epidermidis isolated from postoperative endophthalmitis and the healthy conjunctiva. Sci Rep. 2020;10:14234. doi:10.1038/s41598-020-71050-6
24. Tjaden B. De novo assembly of bacterial transcriptomes from RNA-seq data. Genome Biol. 2015;16:1. doi:10.1186/s13059-014-0572-2
25. Yoon GS, Dong C, Gao N, Kumar A, Standiford TJ, Yu FS. Interferon regulatory factor-1 in flagellin-induced reprogramming: potential protective role of CXCL10 in cornea innate defense against Pseudomonas aeruginosa infection. Invest Ophthalmol Vis Sci. 2013;54:7510–7521. doi:10.1167/iovs.13-12453
26. Li J, Xie L, Qian S, et al. A type VI secretion system facilitates fitness, homeostasis, and competitive advantages for environmental adaptability and efficient nicotine biodegradation. Appl Environ Microbiol. 2021;87(9):e03113–20. doi:10.1128/AEM.03113-20
27. Sheldon JR, Laakso HA, Heinrichs DE. Iron acquisition strategies of bacterial pathogens. Microbiol Spectr. 2016;4. doi:10.1128/microbiolspec.VMBF-0010-2015
28. Evans DJ, Fleiszig SM. Why does the healthy cornea resist Pseudomonas aeruginosa infection? Am J Ophthalmol. 2013;155:961–970, e2. doi:10.1016/j.ajo.2013.03.001
29. Willcox MD. Pseudomonas aeruginosa infection and inflammation during contact lens wear: a review. Optom Vis Sci. 2007;84:273–278. doi:10.1097/OPX.0b013e3180439c3e
30. Naskar S, Hohl M, Tassinari M, Low HH. The structure and mechanism of the bacterial type II secretion system. Mol Microbiol. 2021;115:412–424. doi:10.1111/mmi.14664
31. Alarcon I, Evans DJ, Fleiszig SM. The role of twitching motility in Pseudomonas aeruginosa exit from and translocation of corneal epithelial cells. Invest Ophthalmol Vis Sci. 2009;50:2237–2244. doi:10.1167/iovs.08-2785
32. Piepenbrink KH, Sundberg EJ. Motility and adhesion through type IV pili in Gram-positive bacteria. Biochem Soc Trans. 2016;44:1659–1666. doi:10.1042/BST20160221
33. Burrows LL. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol. 2012;66:493–520. doi:10.1146/annurev-micro-092611-150055
34. Nieto V, Kroken AR, Grosser MR, et al. Type IV pili can mediate bacterial motility within epithelial cells. mBio. 2019;10:e02880–18. doi:10.1128/mBio.02880-18
35. Winstanley C, Kaye SB, Neal TJ, et al. Genotypic and phenotypic characteristics of Pseudomonas aeruginosa isolates associated with ulcerative keratitis. J Med Microbiol. 2005;54:519–526. doi:10.1099/jmm.0.46005-0
36. Thanabalasuriar A, Scott BNV, Peiseler M, et al. Neutrophil extracellular traps confine Pseudomonas aeruginosa ocular biofilms and restrict brain invasion. Cell Host Microbe. 2019;25:526–536, e4. doi:10.1016/j.chom.2019.02.007
37. Sousa AM, Pereira MO. Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs-a review. Pathogens. 2014;3:680–703. doi:10.3390/pathogens3030680
38. Ma L, Wang S, Wang D, Parsek MR, Wozniak DJ. The roles of biofilm matrix polysaccharide Psl in mucoid Pseudomonas aeruginosa biofilms. FEMS Immunol Med Microbiol. 2012;65:377–380. doi:10.1111/j.1574-695X.2012.00934.x
39. Vareechon C, Zmina SE, Karmakar M, Pearlman E, Rietsch A. Pseudomonas aeruginosa effector ExoS inhibits ROS production in human neutrophils. Cell Host Microbe. 2017;21:611–618.e5. doi:10.1016/j.chom.2017.04.001
40. Kaminski A, Gupta KH, Goldufsky JW, Lee HW, Gupta V, Shafikhani SH. Pseudomonas aeruginosa ExoS induces intrinsic apoptosis in target host cells in a manner that is dependent on its GAP domain activity. Sci Rep. 2018;8:14047. doi:10.1038/s41598-018-32491-2
41. Mohankumar V, Ramalingam S, Chidambaranathan GP, Prajna L. Autophagy induced by type III secretion system toxins enhances clearance of Pseudomonas aeruginosa from human corneal epithelial cells. Biochem Biophys Res Commun. 2018;503:1510–1515. doi:10.1016/j.bbrc.2018.07.071
42. Dave A, Samarth A, Karolia R, et al. Characterization of ocular clinical isolates of Pseudomonas aeruginosa from non-contact lens related keratitis patients from South India. Microorganisms. 2020;8:260. doi:10.3390/microorganisms8020260
43. Li W, Lyte M, Freestone PP, Ajmal A, Colmer-Hamood JA, Hamood AN. Norepinephrine represses the expression of toxA and the siderophore genes in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2009;299:100–109. doi:10.1111/j.1574-6968.2009.01739.x
44. Minandri F, Imperi F, Frangipani E, et al. Role of iron uptake systems in Pseudomonas aeruginosa virulence and airway infection. Infect Immun. 2016;84:2324–2335. doi:10.1128/IAI.00098-16
45. Kang D, Kirienko DR, Webster P, Fisher AL, Kirienko NV. Pyoverdine, a siderophore from Pseudomonas aeruginosa, translocates into C. elegans, removes iron, and activates a distinct host response. Virulence. 2018;9:804–817. doi:10.1080/21505594.2018.1449508
46. Wiens JR, Vasil AI, Schurr MJ, Vasil ML. Iron-regulated expression of alginate production, mucoid phenotype, and biofilm formation by Pseudomonas aeruginosa. mBio. 2014;5:e01010–13. doi:10.1128/mBio.01010-13
47. Kang D, Turner KE, Kirienko NV. PqsA promotes pyoverdine production via biofilm formation. Pathogens. 2017;7:3. doi:10.3390/pathogens7010003
48. Molina-Mora JA, García F. Molecular determinants of antibiotic resistance in the Costa Rican Pseudomonas aeruginosa AG1 by a multi-omics approach: a review of 10 years of study. Phenomics. 2021;1:129–142. doi:10.1007/s43657-021-00016-z
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.