Back to Journals » Journal of Inflammation Research » Volume 16
Differential Expression Profiles of Plasma Exosomal microRNAs in Rheumatoid Arthritis
Authors Yang X, Wang Z, Zhang M, Shuai Z
Received 24 March 2023
Accepted for publication 3 August 2023
Published 28 August 2023 Volume 2023:16 Pages 3687—3698
DOI https://doi.org/10.2147/JIR.S413994
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Xiaoke Yang,* Zhixin Wang,* Mingming Zhang, Zongwen Shuai
Department of Rheumatology and Immunology, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zongwen Shuai, Department of Rheumatology and Immunology, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, People’s Republic of China, Tel +86 551 62922247, Fax +86 551 62923040, Email [email protected]
Aim: Differential expression maps of microRNAs (miRNAs) are connected to the autoimmune diseases. This study sought to elucidate the expression maps of exosomal miRNA in plasma of rheumatoid arthritis (RA) patients and their potential clinical significance.
Methods: In the screening phase, small RNA sequencing was performed to characterize dysregulated exosome-derived miRNAs in the plasma samples from six patients with RA and six healthy patients. At the independent verification stage, the candidate plasma exosomal miRNAs were verified in 40 patients with RA and 32 healthy patients by using qRT-PCR. The correlation of miRNA levels and clinical characteristics was tested in patients with RA. The value of these miRNAs in diagnosing RA was assessed with the receiver operating characteristic curve.
Results: During the screening phase, 177 and 129 miRNAs were increased and decreased in RA patients and healthy controls, respectively. There were 10 candidate plasma exosomal miRNAs selected for the next identification. Compared with the healthy controls, eight plasma exosomal miRNAs (let-7a-5p, let-7b-5p, let-7d-5p, let-7f-5p, let-7g-5p, let-7i-5p, miR-128-3p, and miR-25-3p) were significantly elevated in RA patients, but miR-144-3p and miR-15a-5p expression exhibited no significant changes. The let-7a-5p and miR-25-3p levels were linked to the rheumatoid factor-positive phenotype in RA patients. For the eight miRNAs, the area under the subject work characteristic curve (AUC) is 0.641 to 0.843, and their combination had a high diagnostic accuracy for RA (AUC = 0.916).
Conclusion: Our study illustrates that novel exosomal miRNAs in the plasma may represent potential noninvasive biomarkers for RA.
Keywords: rheumatoid arthritis, plasma exosome, microRNAs, biomarker, small RNA sequencing
Introduction
Rheumatoid arthritis (RA) belongs to the autoimmune disease that exhibits chronic systemic symptoms that primarily impair cartilage and bone, leading to joint swelling, pain, impaired movement, and decreased quality of life.1 Approximately 0.5–1.0% of the population are affected by RA worldwide, disproportionately affecting females at a 3:1 ratio to males.2,3 It has been proved that genetic, epigenetic and environmental factors were involved in RA development.4 Many genes have been found to be related to RA susceptibility via genome-wide association studies. Over 80% of the human genome is transcribed with little or no protein-coding capability.5 It was reported that microRNAs (miRNAs) played a key role in the development and progression of autoimmune diseases such as RA, and circulating levels of miRNA may represent potential biomarkers for these diseases.6,7
Cells could secret various extracellular vesicles, including exosomes, and are ubiquitously distributed in bodily fluids.8,9 With the characteristics of stability and biocompatibility, exosomes play essential roles in autoimmune diseases.10 Moreover, exosomes can also serve as carriers through which to contain and deliver a variety of functional substances to assist in intercellular communications.11–13 MiRNAs are defined as single-stranded noncoding RNAs consisting of ~22 nucleotides that participate in regulating post-transcriptional gene expression.14 Therefore, utilizing RNA sequence technology, growing evidence indicates that dysregulated exosomal miRNAs were promising biomarkers for diagnosis and treatment of tumor and autoimmune diseases.15–19 However, there are limited studies investigating the differential expression profiles of miRNAs derived from plasma exosomes in RA.
Exosomes that could be released by various cells have been reported, including inflammatory and immune cells responsible for RA development.20 Growing results suggest that miRNAs contained in exosomes are critically involved in inflammation and immunity.21 Therefore, it is important to identify additional dysregulated miRNAs as novel biomarkers and potential targets for RA therapy. Herein, we investigated expression maps of exosomal miRNAs in plasma of patients with RA versus healthy controls, and analyzed their potential clinical significance.
Materials and Methods
Patients and Data Collection
A two-stage analysis was conducted. For the screening phase, 6 RA patients (1 male and 5 females; 45.17 ± 6.65 years) and 6 healthy age- and sex-matched patients (1 male and 5 females; 40.00 ± 12.63 years) were included. In the validation phase, 40 patients with RA (4 males and 36 females; 47.12 ± 10.72 years) and 32 healthy age- and sex-matched patients (2 males and 30 females; 42.72 ± 10.49 years) were included.
We recruited the patients with RA and healthy patients from the First Affiliated Hospital of Anhui Medical University, and diagnosed in accordance with the American College of Rheumatology (ACR)/European League Against Rheumatism (EULAR) 2010 classification criteria for RA.22 Any RA patients with other autoimmune conditions, serious infections, or pregnancy were eliminated. The healthy patients did not have RA or other autoimmune diseases.
We collected demographic and clinical parameters through questionnaires or medical records. The informed consent was supplied by all participants on the basis of the Helsinki Declaration. The Ethics Committee of Anhui Medical University approved the protocols used in this study.
Extraction of Plasma
Plasma samples were obtained from EDTA-anticoagulant venous blood (10 mL) by centrifugation at 4000 rpm for 10 min and then stored at −80°C in a fresh RNase-free tube preparing for extracting exosomes.
Isolation of Exosomes
In the screening phase, ultra-centrifugation was conducted to isolate exosomes from the plasma samples collected from six RA patients and six healthy controls. During the independent validation phase, we used an exoEasy Maxi Kit (Qiagen, Germany) to extract plasma exosomes from 40 RA patients and 32 healthy controls in accordance with the manufacturer’s protocol. The study subject characteristics in both stages are summarized in Table 1.
 |
Table 1 Clinical Characteristics of RA Patients and Healthy Controls |
Identification of Exosomes
Transmission Electron Microscopy (TEM)
TEM was performed to observe the morphology of exosomes. Briefly, 10 µl diluted exosomes were dripped onto the copper mesh with precipitating for 1 minute. The float was removed by filter paper. Then, 2% phosphotungstic acid was dripped onto the copper mesh with precipitating for 1 minute. After removing the float, the sample was dried at room temperature for a few minutes. The images of exosomes were taken on TEM (HITACHI, HT-7800, Hitachi, Ltd, Japan) at a 80-kV acceleration voltage.
Nano-Flow Cytometry (NanoFCM)
Thirty microliters of diluted exosomes were used to obtain the size and concentration information. The size distribution of isolated exosomes was detected by NanoFCM using High-Sensitivity Flow Cytometry Flow NanoAnalyzer (Fuliu Biotechnology, Ltd, China).
Western Blot (WB)
BCA Protein Assay (Pierce, Thermo Scientific) was used to detect the protein concentrations before WB analysis. Then, the protein samples were configured into 10% gel of sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS PAGE) for electrophoresis. After the proteins were transferred to PVDF membranes, the primary antibody CD9 (1:1000, Abcam, ab92726), CD81 (1:1000, Abcam, ab109201), TSG101 (1:1000, Abcam, ab125011) and Calnexin (1:1000, Abcam, ab22595) were used to incubate proteins at 4°C overnight. Then, the proteins were incubated with secondary antibody. Finally, the proteins were visualized by a chemiluminescence imager (BioRad, China).
Total RNA Extraction
In the small RNA sequencing screening phase, the total RNA was extracted from plasma exosomes with an miRNAeasy Plasma Kit (Qiagen, Germany). In validation phase, total RNA in exosome was abstracted by a miRNAcute miRNA isolation kit (Tiangen Biotech, Beijing, China). All operations were performed in accordance with the manufacturer’s protocol. We used NanoDrop™ 2000 spectrophotometer (Thermo Scientific, USA) to detect the RNA concentrations.
Small RNA Sequencing and Data Analysis
BGI (China) conducted small RNA library preparation, sequencing, and bioinformatic analyses. Library sequencing was carried out with the BGISEQ-500 platform. The sample miRNAs were analyzed using DESeq2 software.
A threshold P value of <0.05 plus a fold change of >1.5 was used to identify the upregulated and downregulated miRNAs. The clustering hierarchy and volcano plot were used to display the differences in exosomal miRNAs expression patterns between the samples. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were carried out to describe the roles of target mRNAs.
miRNA Verification by qRT-PCR
miRcute Plus miRNA First Strand cDNA Synthesis Kit (Tiangen Biotech, Beijing, China) was used to independently verify the data according to the instructions. Subsequently, qRT-PCR assay was conducted with miRcute Plus miRNA qPCR Kit (SYBR Green, Tiangen Biotech, Beijing, China) to measure the candidate miRNAs selected based on the P value and fold change. The following reaction conditions were used: 95°C for 15 min, 5 cycles at 94°C for 20 s, 63°C for 30 s and 72°C for 34 s, followed by 40 cycles at 94°C for 20 s and 60°C for 34 s. All of the primers used for 10 miRNAs (let-7a-5p, let-7b-5p, let-7d-5p, let-7f-5p, let-7g-5p, let-7i-5p, miR-128-3p, miR-25-3p, miR-144-3p, miR-15a-5p) and U6 (as an internal standard) were obtained from Tiangen Biotech. The data were presented as the relative level of target miRNA expression normalized to U6. Each sample was detected in triplicate. Relative quantification of candidate miRNAs expression was according to the 2−ΔΔCt method, with ΔCt = Cttarget − CtU6 and −ΔΔCt = −(sample ΔCt − control ΔCt).
Statistical Analysis
SPSS 23.0 (SPSS Inc., Chicago, IL, USA) software was used to calculate all of the statistical analyses, mean ± SD, or median. The distribution of the continuous variables was described according to the interquartile range. Categorical data was presented as a frequency (percentage). Quantitative data were compared by using Mann–Whitney U-test or Student’s t-test. Mann–Whitney U-test, Student’s t-test, and Spearman's rank correlation analysis were used to analyze the correlation between miRNA expression levels and clinical characteristics of RA. According to the results of univariate analysis and binary logistic regression analysis, the receiver operating characteristic (ROC) curve and the area under the ROC curve (AUC) were performed to indicate the diagnostic power of these exosomal miRNAs. Bilateral P-value <0.05 is statistically significant.
Results
Identification of Plasma Exosomes
TEM revealed that the vesicles were round and uniform in size (Figure 1a). NanoFCM demonstrated that the average particle size was 75.18 nm and the concentration was 6.26E+9 (Particles/mL). Over 99% of particles have diameters between 30 and 150 nanometers (Figure 1b). WB showed that the levels of exosome markers expression (CD9, CD81, TSG101 and Calnexin) could be identified in plasma exosomes (Figure 1c).
 |
Figure 1 Identification of plasma exosomes. (a) TEM photograph of plasma exosomes; (b) NanoFCM of plasma exosomes; (c) results of exosome markers of plasma exosomes by WB. |
Plasma Exosomal miRNA Expression Profiles in RA Patients
According to the SmallRNA sequencing results, there were 177 upregulated and 129 downregulated miRNAs in RA group versus the healthy group. The clustering hierarchy revealed differences in the levels of plasma exosome miRNAs among the samples (Figure 2a). A Volcano plot showed the differentially expressed miRNAs between the different miRNA expression profile groups (Figure 2b).
KEGG Pathway Analysis
Figure 3a and b showed the upregulation and downregulation pathways, respectively. Among the upregulated miRNAs, the MAPK signaling pathway, Rap1 signaling pathway, cGMP-PKG signaling pathway, Wnt signaling pathway and Ras signaling pathway represented the top five enriched signaling pathways. Among the downregulated miRNAs, the MAPK signaling pathway, Rap1 signaling pathway, cGMP-PKG signaling pathway, oxytocin signaling pathway and mTOR signaling pathway is a signal pathway enriched in the top five bits.
 |
Figure 3 KEGG pathway analysis of the differentially expressed exosomal miRNAs. (a) Upregulated; (b) downregulated. |
GO Analysis
The top five meaningful molecular functions were ATP binding, protein binding, nucleotide binding, kinase activity and transferase activity. The top five meaningful cellular components were nucleoplasm, cytosol, nucleus, extracellular exosome and endosome. The top five meaningful biological processes terms were phosphorylation, negative transcription regulation of RNA polymerase II, protein transport, cell cycle and protein phosphorylation (Figure 4).
Level of Candidate Plasma Exosomal miRNAs Validated by qRT-PCR
Using a fold change of >1.5 plus a P value of <0.05 as the selection criteria, 10 candidate miRNAs were selected for further verification in individual samples (Table 2). The 10 miRNAs expression of plasma exosomes from the 40 RA patients and 32 healthy controls were analyzed separately by qRT-PCR during the validation phase. As shown in Table 3 and Figure 5, eight miRNAs were confirmed to be significantly elevated in plasma exosomes of RA patient group compared to the healthy group (all P < 0.05), but two miRNAs exhibited no significant changes (both P > 0.05). Then, RA patients were divided into active and stable RA subgroups. However, there was no significant difference between active RA and stable RA groups (all P > 0.05) (Figure 6).
 |
Table 2 10 Candidate Exosomal miRNAs Detected Using RNA Sequencing |
 |
Table 3 The Expressions of 10 Candidate Exosomal miRNAs in the Preliminary Validation Phase |
 |
Figure 5 (a–j) Comparison of relative levels of 10 plasma exosomal miRNAs between RA patients and control subjects. |
 |
Figure 6 (a–j) Comparison of relative levels of 10 plasma exosomal miRNAs between the stable and active RA patients. |
Correlation Between Significant Plasma Exosomal miRNAs and RA Clinical Parameters
The exosomal let-7a-5p and miR-25a-3p expressed in RA patients were positively associated with RF (Z = 0.363, P = 0.022; Z = 0.317, P = 0.046). However, there was no significant correlation of the eight upregulated exosomal miRNAs with disease duration, DAS28, ESR, CRP and anti-CCP (Table 4). Furthermore, no significant difference in the eight exosomal miRNAs between GC treatment RA group and GC non-treatment RA group was found (all P > 0.05).
 |
Table 4 Correlation Between Clinical Parameters and 8 Upregulated Exosomal miRNAs in RA Patients |
The Diagnostic Power of Significant Plasma Exosomal miRNAs
The AUC values (95% confidence intervals) of the eight exosomal miRNAs elevated in RA patients were 0.843 (0.749, 0.937), 0.762 (0.652, 0.871), 0.774 (0.662, 0.886), 0.752 (0.632, 0.872), 0.825 (0.731, 0.919), 0.641 (0.510, 0.772), 0.702 (0.578, 0.826), and 0.665 (0.536, 0.794), respectively, and that of the eight exosomal miRNAs in combination was 0.916 (0.853, 0.980) (Table 5 and Figure 7).
 |
Table 5 Diagnosis Value of 8 Upregulated Exosomal miRNAs Distinguishing RA Patient and Healthy Subjects |
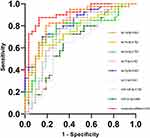 |
Figure 7 ROC curves of eight plasma exosomal miRNAs and combined miRNAs for RA diagnosis. |
Discussion
This study applied small RNA sequencing technology to elucidate a comprehensive miRNA expression map of exosomes in plasma of RA patients and healthy controls. The results revealed that, 306 miRNAs in the plasma exocrine of RA patients were abnormally expressed compared with the healthy control group, including 177 upregulated and 129 downregulated miRNAs. A total of 10 candidate miRNAs were further identified by using RT-qPCR. The levels of eight miRNAs (let-7a-5p, let-7b-5p, let-7d-5p, let-7f-5p, let-7g-5p, let-7i-5p, miR-128-3p, and miR-25-3p) were elevated in plasma exosomes of RA patients, consistent with the small RNA sequencing results. These findings support the reliability of the small RNA sequencing analysis. Additionally, the let-7a-5p and miR-25-3p levels in our study were associated with a rheumatoid factor-positive phenotype in RA patients. Finally, the value of these eight miRNAs was explored as a potential biomarker of RA.
The let-7 family is a novel miRNA, which can regulate the immune killing effect of the immune system by participating in the metabolism, maturation and activation process of immune cells, and is considered to be a new target molecule for immunotherapy.23 Numerous studies have suggested that multiple members of the let-7 family may be involved in the pathogenesis of RA. Recently, a study demonstrated that elevated plasma let-7a-5p could serve as diagnostic marker of RA.24 Another study indicated that let-7a-5p was downregulated in peripheral blood mononuclear cells (PBMCs) of RA.25 Zhu suggested that the expression of let-7a in synovial fluid macrophages was significantly lower in RA patients than in osteoarthritis (OA) patients.26 Besides, let-7a downregulated macrophage activation induced by anti-CCP through targeted binding to HMGA2 and was associated with RA severity. Lai showed that anti-CCP inhibited monocyte let-7a expression in RA patients and promoted inflammation in RA.27 It was reported that the elevated level of exosomal let-7b in the synovial fluid of RA could induce arthritis by combining with Toll-like receptor 7 (TLR-7).28 It has previously been demonstrated that the level of let-7d-5p were increased in the serum and CD8+T cells of RA patients.29,30 The level of let-7f-5p has been identified as a potential biomarker in various autoimmune diseases such as systemic lupus erythematosus (SLE), osteoarthritis (OA), type 1 diabetes (T1D), multiple sclerosis (MS) and myasthenia gravis (MG).31–35 Yang suggested that let-7g-5p was decreased in CD4+T cells and let-7g-5p could alleviate arthritis by inhibiting Th17 cell differentiation.36 Besides, the level of let-7i-5p was increased in the plasma of T1D and the serum of systemic sclerosis (SSc).32,37 The above evidence indicated that the let-7 family is closely associated with the occurrence of multiple autoimmune diseases including RA. Our study is the first study to investigate the association between plasma exosomal let-7 (let-7a-5p, let-7b-5p, let-7d-5p, let-7f-5p, let-7g-5p, let-7i-5p) and RA.
It has been reported that the levels of miR-128-3p were significantly elevated in the plasma, T cells and RA-FLS of RA patients.38–42 Our study first discovered increased level of plasma exosomal miR-128-3p in RA patients. Recently, a bioinformatics analysis showed that miR-25-3p is possibly associated with the regulation of potential RNA regulatory pathway by targeting GZMA in RA.43 Rodríguez-Muguruza found that the serum exosomal miR-25-3p was related to the early diagnosis of RA, which is compatible with our current study results.44 Intriguingly, Rodríguez-Muguruza verified increased levels of miR-144-3p and miR-15a-5p expression in the serum exosomes of early RA patients.44 In contrast, our study revealed no significant differences in exosomal miR-144-3p and miR-15a-5p expression between the RA patients and healthy controls. The reason for this difference may be due to differences sample sizes, methods of measurement, clinical heterogeneity of the patients and the conditions of drug use.
This study was associated with some limitations. First, this study had a relatively small sample size. Therefore, the correlation requires further verification in a larger cohort. Second further studies should be performed to verify whether the changes of these miRNAs in RA patients can be distinguished from other autoimmune diseases. Finally, the detailed mechanism of these miRNAs in plasma exosomes was not explored with respect to the underlying RA development and progression. Thus, additional fundamental studies on miRNAs of interest should be conducted.
The findings of this study reveal the potential value of differentially expressed plasma exosomal miRNA profiles and provide novel insight into the diagnostic biomarkers and promising RA therapeutic targets. However, further experimental studies are necessary to validate the functional mechanism between these identified exosomal miRNAs and RA.
Conclusion
Our study illustrates that novel exosomal miRNAs in the plasma may represent potential noninvasive biomarkers for RA. The let-7a-5p and miR-25-3p levels are correlated with a certain parameter of RA. The potential mechanism between these exosomal miRNAs and RA needs to be verified.
Funding
The current study received support from the Key Project of University Scientific Research Program of the Education Department of Anhui Province (2022AH051173) and the Basic and Clinical Cooperative research Promotion Program of Anhui Medical University (2021xkjT034).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sparks JA. Rheumatoid Arthritis. Ann Intern Med. 2019;170(1):Itc1–itc16. doi:10.7326/AITC201901010
2. Scott DL, Wolfe F, Huizinga TW. Rheumatoid arthritis. Lancet. 2010;376(9746):1094–1108. doi:10.1016/S0140-6736(10)60826-4
3. Favalli EG, Biggioggero M, Crotti C, et al. Sex and management of rheumatoid arthritis. Clin Rev Allergy Immunol. 2019;56(3):333–345. doi:10.1007/s12016-018-8672-5
4. Viatte S, Plant D, Raychaudhuri S. Genetics and epigenetics of rheumatoid arthritis. Nat Rev Rheumatol. 2013;9(3):141–153. doi:10.1038/nrrheum.2012.237
5. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi:10.1038/nature11247
6. Kmiołek T, Paradowska-Gorycka A. miRNAs as biomarkers and possible therapeutic strategies in rheumatoid arthritis. Cells. 2022;11(3):452. doi:10.3390/cells11030452
7. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207. doi:10.1016/j.jaci.2017.08.034
8. Caby MP, Lankar D, Vincendeau-Scherrer C, et al. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17(7):879–87. doi. doi:10.1093/intimm/dxh267
9. Zhang J, Li S, Li L, et al. Exosome and exosomal MicroRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24. doi:10.1016/j.gpb.2015.02.001
10. Xu H, Jia S, Xu H. Potential therapeutic applications of exosomes in different autoimmune diseases. Clin Immunol. 2019;205:116–124. doi:10.1016/j.clim.2019.06.006
11. Kim KM, Abdelmohsen K, Mustapic M, et al. RNA in extracellular vesicles. Wiley Interdiscip Rev RNA. 2017;8(4):e1413. doi:10.1002/wrna.1413
12. Groot M, Lee H. Sorting mechanisms for MicroRNAs into extracellular vesicles and their associated diseases. Cells. 2020;9(4):1044. doi:10.3390/cells9041044
13. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487–514. doi:10.1146/annurev-biochem-013118-111902
14. Bartel DP. Metazoan MicroRNAs. Cell. 2018;173(1):20–51. doi:10.1016/j.cell.2018.03.006
15. Fan Z, Yu J, Lin J, et al. Exosome-specific tumor diagnosis via biomedical analysis of exosome-containing microRNA biomarkers. Analyst. 2019;144(19):5856–5865. doi:10.1039/c9an00777f
16. Hu C, Meiners S, Lukas C, et al. Role of exosomal microRNAs in lung cancer biology and clinical applications. Cell Prolif. 2020;53(6):e12828. doi:10.1111/cpr.12828
17. Mycko MP, Baranzini SE. microRNA and exosome profiling in multiple sclerosis. Mult Scler. 2020;26(5):599–604. doi:10.1177/1352458519879303
18. Mirzaei R, Zamani F, Hajibaba M, et al. The pathogenic, therapeutic and diagnostic role of exosomal microRNA in the autoimmune diseases. J Neuroimmunol. 2021;358:577640. doi:10.1016/j.jneuroim.2021.577640
19. Gong L, Xiao J, Yi J, et al. Immunomodulatory effect of serum exosomes from Crohn disease on macrophages via let-7b-5p/TLR4 signaling. Inflamm Bowel Dis. 2022;28(1):96–108. doi. doi:10.1093/ibd/izab132
20. Hejrati A, Hasani B, Esmaili M, et al. Role of exosome in autoimmunity, with a particular emphasis on rheumatoid arthritis. Int J Rheum Dis. 2021;24(2):159–169. doi:10.1111/1756-185X.14021
21. Duan W, Zhang W, Jia J, et al. Exosomal microRNA in autoimmunity. Cell Mol Immunol. 2019;16(12):932–934. doi:10.1038/s41423-019-0319-9
22. Aletaha D, Neogi T, Silman AJ, et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann Rheum Dis. 2010;69(9):1580–1588. doi:10.1136/ard.2010.138461
23. Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18(10):505–516. doi:10.1016/j.tcb.2008.07.007
24. Liu Y, Han Y, Qu H, et al. Correlation of microRNA expression profile with clinical response to tumor necrosis factor inhibitor in treating rheumatoid arthritis patients: a prospective cohort study. J Clin Lab Anal. 2019;33(7):e22953. doi:10.1002/jcla.22953
25. Tang J, Lin J, Yu Z, et al. Identification of circulating miR-22-3p and let-7a-5p as novel diagnostic biomarkers for rheumatoid arthritis. Clin Exp Rheumatol. 2022;40(1):69–77. doi:10.55563/clinexprheumatol/4me6tg
26. Zhu W, Yu J, Qiu S, et al. MiR-let-7a regulates anti-citrullinated protein antibody-induced macrophage activation and correlates with the development of experimental rheumatoid arthritis. Int Immunopharmacol. 2017;51:40–46. doi:10.1016/j.intimp.2017.08.001
27. Lai NS, Yu HC, Yu CL, et al. Anti-citrullinated protein antibodies suppress let-7a expression in monocytes from patients with rheumatoid arthritis and facilitate the inflammatory responses in rheumatoid arthritis. Immunobiology. 2015;220(12):1351–8. doi. doi:10.1016/j.imbio.2015.07.007
28. Kim S-J, Chen Z, Essani AB, et al. Identification of a novel toll-like receptor 7 endogenous ligand in rheumatoid arthritis synovial fluid that can provoke arthritic joint inflammation. Arthritis Rheumatol. 2016;68(5):1099–1110. doi:10.1002/art.39544
29. Cunningham CC, Wade S, Floudas A, et al. Serum miRNA signature in rheumatoid arthritis and at-risk individuals. Front Immunol. 2021;12:633201. doi:10.3389/fimmu.2021.633201
30. Wasén C, Ospelt C, Camponeschi A, et al. Nicotine changes the microRNA profile to regulate the FOXO memory program of CD8 T cells in rheumatoid arthritis. Front Immunol. 2020;11:1474. doi:10.3389/fimmu.2020.01474
31. Li B, Bai L, Shen P, et al. Identification of differentially expressed microRNAs in knee anterior cruciate ligament tissues surgically removed from patients with osteoarthritis. Int J Mol Med. 2017;40(4):1105–1113. doi:10.3892/ijmm.2017.3086
32. Garavelli S, Bruzzaniti S, Tagliabue E, et al. Blood co-circulating extracellular microRNAs and immune cell subsets associate with type 1 diabetes severity. Int J Mol Sci. 2020;21(2):477. doi:10.3390/ijms21020477
33. Punga T, Bartoccioni E, Lewandowska M, et al. Disease specific enrichment of circulating let-7 family microRNA in MuSK+ myasthenia gravis. J Neuroimmunol. 2016;292:21–26. doi:10.1016/j.jneuroim.2016.01.003
34. Tan W, Gu Z, Leng J, et al. Let-7f-5p ameliorates inflammation by targeting NLRP3 in bone marrow-derived mesenchymal stem cells in patients with systemic lupus erythematosus. Bio Pharmaco. 2019;118:109313. doi:10.1016/j.biopha.2019.109313
35. Z-H L, Wang Y-F, D-D H, et al. Let-7f-5p suppresses Th17 differentiation via targeting STAT3 in multiple sclerosis. Aging. 2019;11(13):4463–4477. doi:10.18632/aging.102093
36. Yang P, Zhang M, Wang X, et al. MicroRNA let-7g-5p alleviates murine collagen-induced arthritis by inhibiting Th17 cell differentiation. Biochem Pharmacol. 2020;174:113822. doi:10.1016/j.bcp.2020.113822
37. Wolska-Gawron K, Bartosińska J, Rusek M, et al. Circulating miRNA-181b-5p, miRNA-223-3p, miRNA-210-3p, let 7i-5p, miRNA-21-5p and miRNA-29a-3p in patients with localized scleroderma as potential biomarkers. Sci Rep. 2020;10(1):20218. doi:10.1038/s41598-020-76995-2
38. Xia Z, Meng F, Liu Y, et al. Decreased MiR-128-3p alleviates the progression of rheumatoid arthritis by up-regulating the expression of TNFAIP3. Biosci Rep. 2018;38(4). doi:10.1042/BSR20180540
39. Peng T, Ji D, Jiang Y. Long non-coding RNA GAS5 suppresses rheumatoid arthritis progression via miR-128-3p/HDAC4 axis. Mol Cell Biochem. 2021;476(6):2491–2501. doi:10.1007/s11010-021-04098-1
40. Murata K, Furu M, Yoshitomi H, et al. Comprehensive microRNA analysis identifies miR-24 and miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS One. 2013;8(7):e69118. doi:10.1371/journal.pone.0069118
41. Wu L, Zhang G, Guo C, et al. MiR-128-3p mediates TNF-α-induced inflammatory responses by regulating Sirt1 expression in bone marrow mesenchymal stem cells. Biochem Biophys Res Commun. 2020;521(1):98–105. doi:10.1016/j.bbrc.2019.10.083
42. Taheri M, Barth DA, Kargl J, et al. Emerging Role of Non-Coding RNAs in Regulation of T-Lymphocyte Function. Front Immunol. 2021;12:756042. doi:10.3389/fimmu.2021.756042
43. Cheng Q, Chen X, Wu H, et al. Three hematologic/immune system-specific expressed genes are considered as the potential biomarkers for the diagnosis of early rheumatoid arthritis through bioinformatics analysis. J Transl Med. 2021;19(1):18. doi:10.1186/s12967-020-02689-y
44. Rodríguez-Muguruza S, Altuna-Coy A, Castro-Oreiro S, et al. A serum biomarker panel of exomiR-451a, exomiR-25-3p and soluble TWEAK for early diagnosis of rheumatoid arthritis. Front Immunol. 2021;12:790880. doi:10.3389/fimmu.2021.790880
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


