Back to Journals » Journal of Inflammation Research » Volume 16
Differential Characteristics of Patients for Hospitalized Severe COVID-19 Infected by the Omicron Variants and Wild Type of SARS-CoV-2 in China
Authors Wei YY , Wang RR, Zhang DW , Chen SH , Tan YY, Zhang WT, Han MF, Fei GH
Received 10 May 2023
Accepted for publication 12 July 2023
Published 21 July 2023 Volume 2023:16 Pages 3063—3078
DOI https://doi.org/10.2147/JIR.S420721
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Yuan-Yuan Wei,1,2,* Rui-Rui Wang,3,* Da-Wei Zhang,1,2,* Su-Hong Chen,2,4 Yuan-Yuan Tan,2,5 Wen-Ting Zhang,1,2 Ming-Feng Han,3 Guang-He Fei1,2
1Department of Respiratory and Critical Care Medicine, First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, 230022, People’s Republic of China; 2Key Laboratory of Respiratory Diseases Research and Medical Transformation of Anhui Province, Hefei, Anhui, 230022, People’s Republic of China; 3Department of Respiratory and Critical Care Medicine, the Second People’s Hospital of Fuyang City, Fuyang, Anhui, 236015, People’s Republic of China; 4Department of Integrated Traditional Chinese and Western Medicine, Anhui Medical University, Hefei, Anhui, 230022, People’s Republic of China; 5Department of Emergency Medicine, First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, 230022, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Guang-He Fei, Department of Respiratory and Critical Care Medicine, First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, 230022, People’s Republic of China, Tel +8655162922013, Fax +8655163635578, Email [email protected] Ming-Feng Han, Department of Respiratory and Critical Care Medicine, the Second People’s Hospital of Fuyang City, Fuyang, Anhui, 236015, People’s Republic of China, Email [email protected]
Background: As multiple mutations of SARS-Cov-2 exist, there are now many viral variants with regional differences in distribution. The clinical characteristics of patients hospitalized with the virus also vary significantly, with those of the Omicron variants being strikingly different from those of the earliest wild-type variant. However, comprehensive data on this subject is lacking. It is therefore crucial to explore these differences to develop better clinical strategies for the management of COVID-19.
Methods: A total of 554 confirmed COVID-19 cases in China were clinically classified as mild, moderate, severe, and critical according to their diagnoses and treatment plans. We compared the demographics and clinical characteristics of patients infected with the Omicron vs wild-type strains, between severe and non-severe cases. Bacterial co-infections with SARS-CoV-2 and correlation between inflammatory factors and T cells were analyzed.
Results: Compared to the wild-type cases, the severe Omicron cases were older (median age 48.36 vs 73.24), and had more upper-respiratory symptoms and comorbidities. Decreased leukocyte counts were less pronounced, although more instances of significantly decreased CD4+ and CD8+ T-cell counts, elevated infection-related biomarkers (eg procalcitonin and C-reactive protein), and abnormal coagulation factors (including increased D-dimer and fibrinogen levels) were detected in the severe Omicron cases. The mean length of hospital stay was significantly shorter in the severe Omicron cases. CD4+ and CD8+ T cell numbers were negatively correlated with neutrophil-to-lymphocyte ratios, as well as serum interleukin-6, procalcitonin, and C-reactive protein levels.
Conclusion: There were significant clinical differences between patients hospitalized with severe cases of Omicron- variant COVID-19 vs wild-type. The Omicron cases tended to be older and had more upper respiratory tract symptoms, comorbidities and bacterial co-infections. Elevated levels of inflammatory cytokines with T-cell depletion correlated with poor disease progression and prognosis. We hope these data provide a theoretical basis for future integrated prevention and control plans for COVID-19.
Keywords: COVID-19, Omicron variants, wild-typeSARS-CoV-2, clinical characteristics, T cell depletion
Introduction
Coronavirus disease 2019 (COVID-19) is an acute fulminant infectious respiratory disease that spread rapidly worldwide.1–3 Human-to-human transmission is the main route of spread for most COVID-19 infections, and its many mutations have raised concerns regarding its pathogenicity and ability to cause severe illness.4 There are now several variants of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) virus, including Alpha, Beta, Gamma, Delta, and Omicron variants, which vary in both temporal and regional distribution. Compared to the wild-type virus, the Omicron variants have several mutations in their spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins and were first identified in South Africa and Botswana on November 24, 2021. The clinical characteristics of current predominant Omicron COVID-19 variants in China are different from those of the earliest epidemic strains, but comprehensive data on the subject is lacking. It is therefore crucial to compare their differences to develop better clinical strategies for their management.
The course of COVID-19 illness can be rapid, causing acute respiratory distress syndrome, septic shock, metabolic acidosis, and dysfunctions in blood coagulation.1,5,6 Although it was announced by the World Health Organization (WHO) that the proportion of severe and critical illnesses related to the Omicron strain decreased as the Omicron BA.2 became the dominant SARS-CoV-2 strain,7–10 there were still large quantities of patients hospitalized with severe COVID-19 as a result of the strain in China, due to its extensive population demographics. The basic reproduction number (R0) of the Omicron variant is between about 10 and 26, suggesting that its transmissibility is quite higher than those of the previous variants. It is therefore essential to identify differences in the clinical characteristics of patients infected with severe cases of Omicron and wild-type COVID-19, to reduce the number of severe cases and reduce the spread of the SARS-CoV-2 virus.
The neutrophil-to-lymphocyte ratio (NLR) is the ratio of the counts of neutrophils and lymphocytes, which serves as an economical biomarker reflecting the inflammatory status of the body and is used in various conditions, including tumors, chronic obstructive pulmonary disease (COPD), and infectious diseases such as influenza and SARS-CoV-2 viral infections.11,12 It has been reported that the NLR has a high capacity to accurately predict the severity of COVID-19.13–15 Lymphocytes are the principal cells involved in the immune response in viral infections. The activation and differentiation of naive T-cells into effector or memory T-cells also play critical roles in antiviral immunity.16 Previous studies have proven that there are low expression levels of angiotensin converting enzyme 2 (ACE2) in T-cells and the SARS-CoV-2 virus may enter this cell type in other way, but cannot replicate further.17–19 However, it has also been reported that T-cell counts, including those of CD4+ and CD8+ cells, are significantly decreased, particularly in cases of severe and critical COVID-19. This can lead to rapid deterioration of the disease, respiratory failure, and even multiple organ failure.1 However, it remains unknown just what causes these decreased T-cell counts following SARS-CoV-2 infection. This study therefore evaluated the clinical characteristics of patients with COVID-19 who were infected with the Omicron variants of the virus, assessed their differences compared to patients who were infected with the wild-type strain, identified the risk factors associated with severe Omicron-related COVID-19, and explored the relationships between T-cells and other inflammatory factors in COVID-19. Through this, we aimed to aid in the development of new strategies for the clinical prevention and treatment of severe COVID-19.
Materials and Methods
Participants
This study complied with the Declaration of Helsinki and was approved by the Ethics Committee of the First Affiliated Hospital of Anhui Medical University (Approval No: 2022371). A total of 554 patients who had been laboratory-confirmed as positive for SARS-CoV-2 infection by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) of oropharyngeal and/or nasopharyngeal swab samples, according to the diagnostic criteria for new coronavirus pneumonia diagnosis and treatment plan (trial version 9)5 from the wards of the First Affiliated Hospital of Anhui Medical University and the Second People’s Hospital of FuYang City. The typing of the viral strains was done in the local laboratory of the Centers for Disease Control, using a Wild-type and Omicron Variant Detection RT-PCR Kit, or was searched through the Global Initiative of Sharing All Influenza Data (GISAID) platform for the genomic epidemiology of SARS-CoV-2 based on the period of the confirmed infection date or the admission date and region (Available online: https://gisaid.org/phylodynamics/global/nextstrain/). There were 401 hospitalized patients infected with the Omicron strain between December 20, 2022, and February 1, 2023; while 153 hospitalized patients were infected with the wild-type strain between January 20, 2020 and February 25, 2020. All patients were clinically classified as mild, moderate, severe, or critical cases, according to the diagnosis and treatment plan.5 There were 116 and 28 severe or critical cases in Omicron and wild-type groups, respectively. All patients involved in this study gave written informed consent to participate.
Data Resources
Epidemiological and clinical data were extracted from electronic medical records. Laboratory parameters were extracted for each patient on their day of admission, including routine blood tests (leucocytes, lymphocyte and neutrophil counts and percentages, hemoglobin and platelets levels), blood biochemistry parameters (albumin, alanine aminotransferase [ALT], aspartate aminotransferase [AST], lactate dehydrogenase [LDH], total bilirubin [Tbil], blood glucose, creatine kinase [CK], creatine kinase isoenzymes [CK-MB], blood urea nitrogen [BUN], total cholesterol, brain natriuretic peptide [BNP] and creatinine clearance [Ccr]), infection-related biomarkers (C-reactive protein [CRP], procalcitonin [PCT] and interleukin-6 [IL-6]) and coagulation function (D-dimer, prothrombin time [PT], activated partial thromboplastin time [APTT] and fibrinogen). NLR was calculated as the ratio of neutrophil-to-lymphocyte counts and PLR was calculated as the ratio of platelet-to-lymphocyte counts. The demographics and clinical characteristics were compared between severe and critical Omicron and wild-type cases, as well as between severe and non-severe Omicron cases. Bacterial co-infections with SARS-CoV-2 and correlations between inflammatory factors and T-cells were also analyzed.
Statistical Analyses
Data analyses were done using SPSS 22.0 software. The Shapiro–Wilk test (S–W test), a type of non-parametric test, was used to determine whether variables conformed to a normal distribution. P values > 0.05 were considered to indicate that the data conformed to a normal distribution. The normally-distributed data were expressed as mean±SD, and compared using the Student’s t-test. The non-normally distributed data were expressed as medians (interquartile ranges) and the rank-sum test was used. Differences between measured data were detected by Chi-squared or the Fisher’s exact tests. When theoretical frequency (T) was ≥ 5 and sample size (N) was ≥ 40, the Chi-squared test was used. When 1 ≤ T < 5 and N ≥ 40, the Chi-squared test with Yates’s correction for continuity was used. When T < 1 or N < 40, Fisher’s exact test was used. Statistical significance in more than two groups was tested using one-way analysis of variance for variables with normal distributions, and the Kruskal–Wallis test otherwise. The correlations between inflammatory factors and T-cell counts were analyzed using Spearson test. P < 0.05 was considered statistically significant.
Results
Comparison of Demographics and Clinical Characteristics Between Hospitalized Patients Infected with Omicron Vs Wild-Type Strain
The mean age of the patients infected with the Omicron strain was 69.00 years, which was significantly higher than the mean age of 40.48 for those infected with the wild-type strain (P < 0.001). There was no statistically significant difference in sex distribution between the Omicron and wild-type groups, at 218 (54.4%) and 86 (56.2%) males, respectively. The incidences of cough, sputum, shortness of breath, chest pain, and muscle ache symptoms were much higher in patients infected with the Omicron strain (all P < 0.001). Of the hospitalized patients in the Omicron group, 84.8% had comorbidities. This was significantly higher than the 26.1% of the wild-type group (P < 0.001; Table 1). The proportion of patients with decreased leucocyte, lymphocyte, and neutrophil counts and percentages was lower in the Omicron group. The levels of infection-related biomarkers, including PCT and CRP, were significantly higher in the Omicron group (P < 0.001). CD4+ and CD8+ T-cell counts were both significantly lower in patients of the Omicron group. The ratio of CD4+ to CD8+ T-cells, however, was not significantly different between the two groups. D-dimer levels were higher, while serum albumin levels were lower, in the Omicron patients. Liver function tests (ALT, AST, Tbil) were normal in most patients. In terms of renal function, BUN levels were higher, but Ccr levels were mostly normal in the Omicron patient group (Table 2).
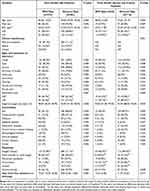 |
Table 1 Comparison of Demographics and Clinical Characteristics Between Hospitalized Patients Infected with Wild and Omicron Strains |
 |
Table 2 Comparison of Laboratory and Radiographic Results Between Hospitalized Patients Infected with Wild and Omicron Strains |
Comparison of Demographics and Clinical Characteristics Between Severe and Critical Omicron and Wild-Type Cases
To further explore differences in the treatment strategies for severe COVID-19 cases, we compared the characteristics between hospitalized severe and critical with Omicron (116 cases) and wild-type (28 cases). The mean age was 73.24 in the severe/critical Omicron group, which was significantly higher than 48.36 in the severe/critical wild-type group (P < 0.001). The percentages of patients with upper-respiratory symptoms, including cough, sputum, and shortness of breath; and the proportions of comorbidities such as cardiovascular, respiratory, and central nervous system diseases, were significantly higher in the Omicron group (all P values < 0.05; Table 1). Decreased leucocyte, total lymphocyte, and neutrophil counts were less pronounced in severe/critical patients of the Omicron, compared to the wild-type, although CD4+ and CD8+ T-cell counts were significantly lower in patients infected with the Omicron variant (P = 0.002 and P = 0.004, respectively). The percentage of severe cases with increased levels of IL-6 was similarly high between the different strain groups (P = 0.246), but that of infection-related biomarkers, including PCT and CRP (as indicators of bacterial co-infections), were significantly higher in the severe Omicron group (P < 0.001). Abnormal coagulation, including increased D-dimer and fibrinogen levels, was significantly higher in the severe and critical Omicron groups (both P values < 0.001). Serum albumin levels were significantly lower in severe and critical Omicron groups (P < 0.001). The results of liver function tests, including AST and Tbil were similar between the groups, but biomarkers of myocardial damage, including CK-MB and LDH, as well as levels of the renal function indicator BUN, were significantly higher in the severe and critical Omicron groups. Ccr levels were 18.1% lower in the 116 severe Omicron cases, and 21.4% lower in the wild-type cases, with no significant difference (P = 0.686; Table 2). Most cases in both strain groups required oxygen therapy, and the rate of use of invasive mechanical ventilation was higher for the severe and critical Omicron groups. The length of hospital stay were significantly lower in severe and critical Omicron groups (P = 0.017; Table 1).
Comparisons of Clinical Characteristics Between Severe and Non-Severe Patients with Omicron-Variant COVID-19
According to our clinical classification criteria, the 285 patients with mild or moderate Omicron-variant COVID-19 were classified into the non-severe group, and the other 116 with severe or critical cases of the disease were placed into the severe group, for the purposes of this study. The mean age of the patients in the severe group was 73.24, while that of the non-severe patients was 65.00 years, which was statistically different (P < 0.001). Compared to the non-severe patients, there were significantly more comorbidities, including cardiovascular and central nervous system diseases, in the severe group (P = 0.012 and P = 0.001, respectively; Table 3). Compared to the non-severe group, CD4+ and CD8+ T-cell and serum albumin levels were significantly decreased, while neutrophil counts and levels of BNP, CK, CK-MB, LDH, PCT, CRP, IL-6, and NLR were significantly higher in the severe group (all P values < 0.05). Increased D-dimer, PT, APTT, and fibrinogen levels, which suggested abnormal coagulation, were found in the severe and critical Omicron groups (P < 0.001). The percentage of patients with increased fasting blood glucose levels was 70.7% in the severe Omicron group, which was significantly higher than the 40.4% of the non-severe Omicron group (P < 0.001; Table 4). Compared to the non-severe group, the proportion of patients with bacterial co-infections was significantly higher in the severe and critical Omicron groups. Older patients, as well as those who had obesity, decreased lymphocyte counts, increased inflammatory marker and blood glucose levels, abnormal coagulation functions, and bacterial co-infections, were found to be at higher risk of developing severe Omicron-variant COVID-19.
 |
Table 3 Demographics and Clinical Characteristics of Hospitalized Patients with Omicron Variant |
 |
Table 4 Laboratory and Radiographic Results of Patients with Omicron Variant |
Bacterial Co-Infections
Compared to the non-severe patients, the proportion of patients who had bacterial co-infections was significantly higher in the severe and critical Omicron groups. In the non-severe patients, the detection rates for Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, Staphylococcus aureus, Escherichia coli, and other bacteria were 30.77%, 15.38%, 23.08%, 7.69%, 7.69%, and 15.38%, respectively. In severe patients, the detection rates of Acinetobacter baumannii, Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus aureus, Escherichia coli, and other bacteria were 47.83%, 26.09%, 8.70%, 4.35%, 4.35%, and 8.70%, respectively (Figure 1).
Correlation Analysis Between Inflammatory Factors and T-Cells
Serum levels of IL-6, CRP, PCT, and NLR, as well as CD4+ and CD8+ T-cell counts, were evaluated for the patients with severe COVID-19, and the results revealed that elevated levels of IL-6, CRP, PCT, and NLR were negatively correlated with both CD4+ and CD8+ counts in cases of severe or critical COVID-19. This suggested that increased levels of inflammatory cytokines may lead to T-cell depletion. The correlation coefficients between CD4+ counts and levels of IL-6, CRP, PCT, and NLR were 0.2223, 0.2732, 0.3512, and 0.5213, respectively (P = 0.0262, P = 0.0018, P < 0.0001 and P < 0.0001, respectively). The correlation coefficients between CD8+ counts and levels of IL-6, CRP, PCT, and NLR were 0.2189, 0.2559, 0.3440, and 0.5177 (P = 0.0287, P = 0.0035, P < 0.0001 and P < 0.0001; Figure 2).
Discussion
At the time of writing this report, the Omicron variant of the SARS-CoV-2 virus has a particularly high rate of transmission and has spread rapidly. The sharp increase in patients hospitalized with severe Omicron COVID-19 has placed great pressure on medical institutions across the country. Although the overall pathogenicity of the Omicron variant has decreased in some countries,7–9,20,21 the rate of patients who develop severe diseases in China, which has a particularly large population, cannot be ignored. Omicron variants are constantly emerging, and our understanding regarding the differences in clinical characteristics of patients that are caused by the different variants is still very limited. This study summarized the differences in the clinical characteristics between the Omicron and wild-type variants, the risk factors related to developing severe Omicron-related disease, and explored the relationships between T-cell counts and inflammatory factors in COVID-19, to provide more evidence for the development of future prevention and treatment strategies for the disease.
In this study, the percentage of male patients was comparable to females, which indicated there were no sex-based differences between Omicron and wild-type COVID-19. The mean age of hospitalized patients infected with the Omicron strain was significantly higher than that of patients infected with the wild strain, and even higher in those who had severe Omicron infections. This indicated that the elderly are more susceptible to the Omicron variant, and are more likely to develop severe COVID-19 from it, consistent with the results of previous studies in Japan and South Africa.22–24
Compared to previous variants, upper-respiratory symptoms, including cough, sputum, shortness of breath, and others, were more frequently observed in Omicron COVID-19. Previous studies have suggested that the viral load levels in the noses of hamsters infected with the Omicron strain were similar to those of animals infected with the earlier strains, but that the levels in their lungs were 10% lower.25 The TMPRSS2 proteins that help most strains of SARS-CoV-2 enter cells are highly expressed on the surfaces of many lung cells but have low levels of expression in the upper-respiratory tract.26,27 The Omicron variant does not bind to TMPRSS2 as well as some previous strains of the virus and preferentially enters cells in the upper-respiratory tract through other routes such as endocytosis, after which it also replicates faster than the earlier viral strains.27,28 This indicates a higher transmissibility of the strain, and that positivity and accuracy rates of nucleic acid sampling are highest for samples taken from the nasopharynx, for the Omicron strain.
In this study, liver function tests, including AST and Tbil, and biomarkers related to myocardial damage, including CK, CK-MB, and LDH, as well as levels of the renal function indicator BUN, were significantly elevated in severe cases of both Omicron and wild-type COVID-19, suggesting that COVID-19 affects multiple organs. It should be noted that the percentage of increased biomarkers related to myocardial damage was as high as 8% in cases of severe Omicron COVID-19, and some patients with mild diseases experienced sudden death even though their clinical symptoms were minor or had improved significantly. It has been reported that extensive impairments of left ventricular systolic function in the early stages of COVID-19 were associated with the severity of the infection.29,30 It has been speculated that SARS-CoV-2 may attack the heart by activating ACE2 in endothelial cells of the cardiovascular system and that an inflammatory storm may play a key role in the progression of the disease, but this notion merits further exploration.31 Many patients with severe COVID-19 in this study had comorbidities, including cardiovascular, digestive, and respiratory diseases, as well as diabetes. This proportion was much higher in the group that had severe Omicron-related COVID-19. Chronic respiratory diseases such as COPD may lead to chronic hypoxia and decreased oxygen saturation, which predisposes patients to severe Omicron COVID-19. Decreased fingertip oxygen saturation is a risk factor for severe disease; therefore, providing suitable oxygen therapy and adequate oxygen supply for patients is important. In our study, 70.7% of the patients had hyperglycemia and 27.6% of patients with severe Omicron COVID-19 had diabetes, which can inhibit the phagocytosis of white blood cells and worsen immune function.32 During the SARS outbreak, diabetic patients had a 3× higher rate of mortality, admission to intensive care units (ICUs), and treatments that required mechanical ventilation.33 One study published in the Lancet suggested that medications used to treat diabetes, such as metformin, glitazone, and betel, may prevent or suppress acute respiratory distress syndrome (ARDS) and reduce mortality.34 Therefore, glucose levels should be monitored and adjusted continually for patients with COVID-19, especially for diabetic ones, to reduce the risk of disease progression.
It has been reported that the levels of inflammatory reactions in cases of Omicron infection were not as severe as those of Delta infection, However, in this study, the levels of inflammatory mediators and cytokines including CRP, IL-6, fibrinogen, and NLR, were higher in patients with severe Omicron COVID-19, which may reflect excessive activation of the immune system.35 It is therefore still necessary to adopt appropriate anti-inflammatory therapies at the most optimal times. Glucocorticoids have been recommended for severe or critical cases with worsening oxygenation, rapid imaging progression, or the activation of excessive inflammatory responses. However, it is critically important to carefully control the occasion, dose, and course of this type of therapy. When the immune system is exhausted, particularly in cases of severe COVID-19 where patients require endotracheal intubation, glucocorticoid treatments can prolong the time taken for virus clearance and lead to secondary infections.36 Immune cell and inflammatory factor levels typically return to normal, however, in discharged patients. Therefore, inflammatory factors, including CRP, IL-6, fibrinogen, and NLR can serve as effective biomarkers to predict disease prognosis and direct the use of anti-inflammatory therapy in cases of severe COVID-19.
This study demonstrated that decreased counts of leucocytes, lymphocytes, and neutrophils were less pronounced in severe and critical patients with Omicron COVID-19 compared to those with the wild-type disease, but that CD4+ and CD8+ T-cell counts were significantly decreased. Decreased CD4+ and CD8+ levels were found to be risk factors for severe COVID-19. The activation and differentiation of T-cells play critical roles in antiviral immunity by secreting various cytokines,37 and there have been reports of inflammatory storms accompanied by the production of excessive inflammatory factors in cases of severe Omicron COVID-19.38 Our results showed that elevated levels of inflammatory factors were negatively correlated with both CD4+ and CD8+ counts in cases of severe and critical COVID-19, suggesting that excessive inflammatory responses contributed to the depletions of T cell, and in turn led to rapid deterioration of the disease, respiratory failure, and even multiple organ failure. T-cell exhaustion also aggravated immune system imbalances, and ultimately caused poor prognosis.39,40 Therefore, the rapid decline of T-cell counts can be a clinical warning indicating severe Omicron infection. Treatments based on inhibiting T-cell exhaustion and promoting the differentiation of T-cells into long-term memory T-cells to quickly respond to and fight the virus, as well as avoid re-infection,40 may represent a future direction for the treatment of severe COVID-19 infection.
In this study, there were high percentages of bacterial co-infections in patients who were hospitalized for severe Omicron COVID-19. Furthermore, the bacterial spectrum of co-infection varies greatly among patients with different disease severities. Klebsiella pneumoniae and Pseudomonas aeruginosa accounted for the highest proportion in patients with non-severe diseases, while Acinetobacter baumannii was detected the most in patients with severe COVID-19. These results differ from others, which reported that the detection rates of Klebsiella pneumoniae and Aspergillus fumigatus in patients with severe COVID-19 were higher than those in patients with non-severe disease in Guangzhou, China.41 This discrepancy may be due to regional differences. Meanwhile, temperature, neutrophil counts, and levels of PCT and CRP were found to be significantly increased in cases of severe Omicron COVID-19, which may indicate bacterial co-infection. Therefore, it is necessary to strengthen the monitoring of co-infections in patients with severe COVID-19 and avoid inappropriate treatments involving antibiotics, particularly broad-spectrum ones. Rapid and accurate etiological diagnoses should be performed, to provide solid laboratory bases for the precise usage of antibiotics in cases of severe Omicron COVID-19.
The major limitation of this study is its retrospective design and lack of follow-up data. The duration of the study was also insufficient to classify results according to the subtypes of Omicron variants, which may have impacted the study’s ability to provide comprehensive details regarding the differential characteristics between the Omicron variant and wild-type COVID-19, in terms of guiding clinical treatments. Therefore, prospective multicenter studies with larger sample studies are warranted to further explore this topic in the future.
Conclusions
The Omicron variant of the SARS-CoV-2 virus has a particularly high rate of transmission and has spread rapidly. The sharp increase in patients hospitalized with severe Omicron COVID-19 has placed great pressure on medical institutions across the country. Omicron variants are constantly emerging, and it is crucial to investigate the differences in clinical characteristics of patients that are caused by the different variants. This study proved that there were significant clinical differences between patients hospitalized with severe cases of Omicron-variant COVID-19 vs wild-type. The Omicron cases tended to be older and had more upper respiratory tract symptoms, comorbidities and bacterial co-infections. Elevated levels of inflammatory cytokines with T-cell depletion correlated with poor disease progression and prognosis. These data provide a theoretical basis for future integrated prevention and control plans for COVID-19.
Abbreviations
SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; ACE2, angiotensin converting enzyme 2; RT-PCR, Reverse Transcription-Polymerase Chain Reaction; PCT, procalcitonin; CRP, C-reactive protein; ALT, Alanine aminotransferase; AST, Aspartate aminotransferase; Tbil, total bilirubin; BUN, blood urea nitrogen; Ccr, Creatinine Clearance Rate; IL-6, interleukin 6; CK, creatine kinase; CK-MB, isoenzyme of creatine kinase; LDH, lactate dehydrogenase; NLR, neutrophil to lymphocyte ratio; PT, Prothrombin time; APTT, activated partial thromboplastin time; SARS, Severe Acute Respiratory Syndrome; ICU, Intensive Care Unit; ARDS, acute respiratory distress syndrome.
Data Sharing Statement
All data reported in this study are available from the corresponding authors (Guang-He Fei and Ming-Feng Han) upon reasonable request, following institutional approval.
Ethics Approval
The study complied with the Declaration of Helsinki and was approved by the First Affiliated Hospital of Anhui Medical University Ethics Committee (No: 2022371).
Consent for Publication
All patients involved in this study gave written informed consent to participate, and the authors have reviewed and approved the final version of the manuscript.
Author Contributions
All authors made a significant contributions to the work reported, including the conception, study design, execution, acquisition of data, analysis and interpretation, in addition to participating in drafting, revising, or critically reviewing the article. All authors provided final approvals of the version to be submitted for publication, as well as agreed on the journal to which the article has been submitted, with full accountability for all aspects of the work.
Funding
This work was supported by a grant from the Key Technology Research and Development Program of Anhui Province (No. 2022e07020076).
Disclosure
The authors have no conflicts of interest to disclose for this work.
References
1. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi:10.1016/S0140-6736(20)30183-5
2. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi:10.1056/NEJMoa2001017
3. Liu X, Cao J, Ji Y, et al. An innovative two-wing model for balancing the demands of inpatients with COVID-19 and general medical service in a designated hospital for COVID-19 in Shenzhen, China. Biosci Trends. 2022;16(2):163–166. doi:10.5582/bst.2022.01106
4. Phan LT, Nguyen TV, Luong QC, et al. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020;382(9):872–874. doi:10.1056/NEJMc2001272
5. Diagnosis and treatment of pneumonia caused by 2019-nCoV (version 9); 2020. Available from: http://www.nhc.gov.cn/yzygj/s7653p/202203/b74ade1ba4494583805a3d2e40093d88.shtml.
6. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi:10.1016/S0140-6736(20)30211-7
7. Lewnard JA, Hong VX, Patel MM, Kahn R, Lipsitch M, Tartof SY. Clinical outcomes associated with SARS-CoV-2 Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in Southern California. Nat Med. 2022;28(9):1933–1943. doi:10.1038/s41591-022-01887-z
8. Skarbinski J, Wood MS, Chervo TC, et al. Risk of severe clinical outcomes among persons with SARS-CoV-2 infection with differing levels of vaccination during widespread Omicron (B.1.1.529) and Delta (B.1.617.2) variant circulation in Northern California: a retrospective cohort study. Lancet Reg Health Am. 2022;12:100297. doi:10.1016/j.lana.2022.100297
9. Veneti L, Bøås H, Bråthen Kristoffersen A, et al. Reduced risk of hospitalisation among reported COVID-19 cases infected with the SARS-CoV-2 Omicron BA.1 variant compared with the Delta variant, Norway, December 2021 to January 2022. Eurosurveillance. 2022;27(4):2200077. doi:10.2807/1560-7917.ES.2022.27.4.2200077
10. Chu J, Morikawa H, Chen Y. Simulation of SARS-CoV-2 epidemic trends in Tokyo considering vaccinations, virus mutations, government policies and PCR tests. Biosci Trends. 2023;17:38–53. doi:10.5582/bst.2023.01012
11. Parthasarathi A, Padukudru S, Arunachal S, et al. The role of neutrophil-to-lymphocyte ratio in risk stratification and prognostication of COVID-19: a systematic review and meta-analysis. Vaccines. 2022;10(8):1233. doi:10.3390/vaccines10081233
12. Russell CD, Parajuli A, Gale HJ, et al. The utility of peripheral blood leucocyte ratios as biomarkers in infectious diseases: a systematic review and meta-analysis. J Infect. 2019;78(5):339–348. doi:10.1016/j.jinf.2019.02.006
13. Sayed AA, Allam AA, Sayed AI, Alraey MA, Joseph MV. The use of neutrophil-to-lymphocyte ratio (NLR) as a marker for COVID-19 infection in Saudi Arabia: a case-control retrospective multicenter study. Saudi Med J. 2021;42(4):370–376. doi:10.15537/smj.2021.42.4.20200818
14. Wang Y, Zhao J, Yang L, Hu J, Yao Y. Value of the neutrophil-lymphocyte ratio in predicting COVID-19 severity: a meta-analysis. Dis Markers. 2021;2021:2571912. doi:10.1155/2021/2571912
15. Sarkar S, Khanna P, Singh AK. The impact of neutrophil-lymphocyte count ratio in COVID-19: a systematic review and meta-analysis. J Intensive Care Med. 2022;37(7):857–869. doi:10.1177/08850666211045626
16. Peng Y, Mentzer AJ, Liu G, et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21(11):1336–1345. doi:10.1038/s41590-020-0782-6
17. Wang Q, Zhang Y, Wu L, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904.e9.
18. Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS-CoV-2 Receptor ACE2 Is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181(5):1016–35.e19. doi:10.1016/j.cell.2020.04.035
19. Dong M, Zhang J, Ma X, et al. ACE2, TMPRSS2 distribution and extrapulmonary organ injury in patients with COVID-19. Biomed Pharmacother. 2020;131:110678. doi:10.1016/j.biopha.2020.110678
20. Menni C, Valdes AM, Polidori L, et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: a prospective observational study from the ZOE COVID Study. Lancet. 2022;399(10335):1618–1624. doi:10.1016/S0140-6736(22)00327-0
21. Abdullah F, Myers J, Basu D, et al. Decreased severity of disease during the first global omicron variant covid-19 outbreak in a large hospital in Tshwane, South Africa. Int J Infect Dis. 2022;116:38–42. doi:10.1016/j.ijid.2021.12.357
22. Wolter N, Jassat W, Walaza S, et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet. 2022;399(10323):437–446. doi:10.1016/S0140-6736(22)00017-4
23. Maslo C, Friedland R, Toubkin M, Laubscher A, Akaloo T, Kama B. Characteristics and outcomes of hospitalized patients in South Africa during the COVID-19 Omicron wave compared with previous waves. JAMA. 2022;327(6):583–584. doi:10.1001/jama.2021.24868
24. Morioka S, Tsuzuki S, Suzuki M, et al. Post COVID-19 condition of the Omicron variant of SARS-CoV-2. J Infect Chemother. 2022;28(11):1546–1551. doi:10.1016/j.jiac.2022.08.007
25. Halfmann PJ, Iida S, Iwatsuki-Horimoto K, et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603(7902):687–692. doi:10.1038/s41586-022-04441-6
26. Hui KPY, Ng KC, Ho JC, et al. Replication of SARS-CoV-2 Omicron BA.2 variant in ex vivo cultures of the human upper and lower respiratory tract. EBioMedicine. 2022;83:104232. doi:10.1016/j.ebiom.2022.104232
27. Hui KPY, Ho JC, Cheung MC, et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature. 2022;603(7902):715–720. doi:10.1038/s41586-022-04479-6
28. Fan Y, Li X, Zhang L, Wan S, Zhang L, Zhou F. SARS-CoV-2 Omicron variant: recent progress and future perspectives. Signal Transduct Target Ther. 2022;7(1):141. doi:10.1038/s41392-022-00997-x
29. Wan EYF, Mathur S, Zhang R, et al. Association of COVID-19 with short- and long-term risk of cardiovascular disease and mortality: a prospective cohort in UK Biobank. Cardiovasc Res. 2023;119:1718–1727. doi:10.1093/cvr/cvac195
30. Li R, Wang H, Ma F, et al. Widespread myocardial dysfunction in COVID-19 patients detected by myocardial strain imaging using 2-D speckle-tracking echocardiography. Acta Pharmacol Sin. 2021;42(10):1567–1574. doi:10.1038/s41401-020-00595-z
31. Patone M, Mei XW, Handunnetthi L, et al. Risks of myocarditis, pericarditis, and cardiac arrhythmias associated with COVID-19 vaccination or SARS-CoV-2 infection. Nat Med. 2022;28(2):410–422. doi:10.1038/s41591-021-01630-0
32. Yang JK, Lin SS, Ji XJ, Guo LM. Binding of SARS coronavirus to its receptor damages islets and causes acute diabetes. Acta Diabetol. 2010;47(3):193–199. doi:10.1007/s00592-009-0109-4
33. Booth CM, Matukas LM, Tomlinson GA, et al. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA. 2003;289(21):2801–2809. doi:10.1001/jama.289.21.JOC30885
34. Zumla A, Hui DS, Azhar EI, Memish ZA, Maeurer M. Reducing mortality from 2019-nCoV: host-directed therapies should be an option. Lancet. 2020;395(10224):e35–e6. doi:10.1016/S0140-6736(20)30305-6
35. Guo XJ, Thomas PG. New fronts emerge in the influenza cytokine storm. Semin Immunopathol. 2017;39(5):541–550. doi:10.1007/s00281-017-0636-y
36. Vandewalle J, Luypaert A, De Bosscher K, Libert C. Therapeutic Mechanisms of Glucocorticoids. Trends Endocrinol Metab. 2018;29(1):42–54. doi:10.1016/j.tem.2017.10.010
37. Dong C. Cytokine regulation and function in T cells. Annu Rev Immunol. 2021;39:51–76. doi:10.1146/annurev-immunol-061020-053702
38. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected Pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi:10.1001/jama.2020.1585
39. Khan O, Giles JR, McDonald S, et al. TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature. 2019;571(7764):211–218. doi:10.1038/s41586-019-1325-x
40. Alfei F, Kanev K, Hofmann M, et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature. 2019;571(7764):265–269. doi:10.1038/s41586-019-1326-9
41. Cheng LL, Li SY, Zhong NS. 应注意当前重症新型冠状病毒感染的新特点 [New characteristics of COVID-19 caused by the Omicron variant in Guangzhou]. Chin J Tubercul Res Dis. 2023;46(5):441–443. Chinese. doi:10.3760/cma.j.cn112147-20230311-00125
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


