Back to Journals » Infection and Drug Resistance » Volume 16
Diagnostic Efficacy of Metagenomic Next-Generation Sequencing in Patients with Spinal Infections: A Retrospective Study
Authors Cheng H , Wu H, Tan N, Liu Z, Wang N, Chen N, Li C
Received 15 September 2023
Accepted for publication 6 December 2023
Published 12 December 2023 Volume 2023:16 Pages 7613—7620
DOI https://doi.org/10.2147/IDR.S435466
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Hanwen Cheng,1,* Haoyu Wu,1,* Ni Tan,2,* Zhuojie Liu,1 Ning Wang,1 Ningyi Chen,1 Chunhai Li1,3
1Department of Orthopaedics, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, Guangdong Province, People’s Republic of China; 2Cellular and Molecular Diagnostics Center, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou, Guangdong Province, People’s Republic of China; 3Teaching and Research Bureau of Surgery, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, Guangdong Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chunhai Li, Department of Orthopaedics, Sun Yat-sen Memorial Hospital, Sun Yat‐sen University, Guangzhou, Guangdong Province, 510000, People’s Republic of China, Tel +86-18602079796, Fax +86-(20)81332523, Email [email protected]
Purpose: Early diagnosis of spinal infections remains challenging, and emerging metagenomic next-generation sequencing (mNGS) technology holds promise in addressing this issue. The aim of this study is to investigate the diagnostic efficacy of mNGS in spinal infections.
Patients and Methods: A total of 78 cases with suspected spinal infections were enrolled in this study, all of whom underwent laboratory, histopathological and mNGS examinations upon admission. Lesion samples were obtained by surgical or C-arm-guided puncture. Sensitivity, specificity, positive predictive value and negative predictive value of culture and mNGS were calculated for statistical analysis.
Results: With histopathological results as the reference, the included 78 patients were categorized into 50 cases in the spinal infection group and 28 cases in the aseptic group. The sensitivity (84%) and negative predictive value (77.14%) of mNGS were significantly higher than those of culture (32% and 44.26%, respectively), whereas no significant differences were observed in terms of specificity and positive predictive value. In the subgroup analysis for Mycobacterium tuberculosis, the sensitivity of mNGS (90.91%) and T-spot (90.91%) was significantly higher than that of culture (0). Additionally, mNGS demonstrated markedly higher specificity (100%) compared to T-spot (85.07%).
Conclusion: This study underscores the substantial advantages of mNGS in terms of diagnostic accuracy and bacterial coverage for spinal infections. The findings provide compelling clinical evidence that supports the enhanced diagnostic efficacy of mNGS.
Keywords: culture, diagnosis, sensitivity, specificity, bacteria, clinical evidence
Introduction
Spinal infection, an uncommon condition, was first recorded in 1779.1 When the intervertebral disk is infected, it is commonly referred to as spondylodiscitis,2 whereas infection of the vertebral body or endplates is more precisely termed vertebral osteomyelitis or spondylitis.3 The definitive diagnosis of spinal infection is often delayed by 2–6 months due to the atypical clinical signs and the low diagnostic efficiency of traditional cultivation methods.4 Consequently, the timely initiation of antibiotic treatment is often missed, and significant structural damage to the spine is frequently already present by the time a definitive diagnosis is made.
As a novel high-throughput sequencing method, metagenomic next-generation sequencing (mNGS) has been extensively employed for identifying the infecting organism, particularly microorganisms that are challenging to culture.5 Given that the mNGS technique enables rapid and impartial culture-independent diagnosis, it has been adopted in clinical laboratories for pneumonia, sepsis, and spinal infection.6–8 In current clinical practice, numerous case reports and retrospective studies have demonstrated the superior diagnostic efficiency of mNGS compared to traditional culture methods.9–11 Moreover, the use of antibiotics significantly undermines the success of traditional cultures, whereas the mNGS technique is minimally affected.12 Nevertheless, there remains a scarcity of studies evaluating the accuracy and validity of mNGS in identifying infectious organisms in spinal infections.
In recent years, with the declining cost of sequencing, mNGS has been implemented in clinical practice, offering substantial benefits to patients with suspected spinal infections.7 Herein, a retrospective study was conducted to evaluate the accuracy and validity of mNGS in diagnosing spinal infections.
Materials and Methods
Participants
This retrospective study encompassed patients with suspected spinal infections who sought treatment at Sun Yat-sen memorial hospital from January 2021 to July 2023. Patients with suspected spinal infections were identified based on their clinical symptoms, laboratory tests, and imaging.1,13 All the cases included underwent both histopathological examination and mNGS.
Methods of Sample Acquisition
Upon admission, we adhered to the guidelines outlined by the Infectious Diseases Society of America (IDSA) for diagnosing and treating native vertebral osteomyelitis in adults. Patients meeting criteria for surgical intervention underwent relevant procedures to acquire lesion samples. Conversely, for those not meeting surgical criteria, we followed the guidelines, employing a puncture method to obtain samples from the lesion. These interventions were carried out in a standard orthopedic operating room and were performed by a highly experienced spine surgeon with over thirty years of clinical practice.
Laboratory Tests
All patients fast for 8 to 12 hours after admission and undergo fasting venous blood collection on the following morning for subsequent testing. 1) The Ottoman-1000 fully automated specific protein detection analyzer was utilized to measure C-reactive protein (CRP) levels in whole blood using the latex-enhanced immunoturbidimetric method. A CRP level exceeding 5 mg/L was considered positive. 2) The Westergren method was used to measure the erythrocyte sedimentation rate (ESR). 3) The white blood cell (WBC) testing was performed using the Sysmex XS-1000i fully automated hematology analyzer along with its original reagents. A WBC count exceeding 10×10^9/L was considered positive. 4) We separate peripheral blood mononuclear cells (PBMCs) from venous blood and utilize the T-SPOT.TB assay kit (Oxford Immu-Notec Co., LTD, UK) for detection. Interpretation of results: when the spot count in the negative control wells is between 0–5, if the count of spots in any well containing CFP-10 or ESAT-6 antigens minus the count in the negative control well is ≥6, it is considered positive. If the count in the negative control wells is ≥6, and the count in any detection well is more than 2 times the count in the negative control wells, it is deemed positive. In cases where the count in the positive control well is ≥20, and the count in the detection wells does not meet the positive criteria, it is considered negative.
Aerobic, Anaerobic and Fungal Cultures
If a patient was suspected of having a spinal infection, blood samples were collected first for aerobic, anaerobic, and fungal cultures upon the patient’s admission to the hospital. Subsequently, upon obtaining samples from the lesion site, they are divided into multiple parts for culture testing. Aerobic and fungal samples were stored in sterile test tubes, while sterile test tubes filled with saline were used to store anaerobic samples. The cultivation period for all bacteria was set at 7 days, and if no positive results were observed within this time frame, it was determined as negative.
Histopathologic Examination
Fresh samples were swiftly transported to the pathology department of Sun Yat-sen Memorial Hospital in sterile test tubes. As per previously published research, histopathological results were used as the criteria in this study to determine whether a patient falls into the category of spinal infection.1,10
Metagenomic Next-Generation Sequencing
The samples were stored in a sterile container, then preserved and transported to Cellular & Molecular Diagnostics Center of Sun Yat-sen memorial hospital within 30 minutes in liquid nitrogen. The whole process of sequencing and pathogen detection pipeline were carried out in the laboratory of Cellular & Molecular Diagnostics Center of Sun Yat-sen memorial hospital, Sun Yat-sen university, Guangzhou, China. Further comprehensive details can be found in the Supplementary Material.
Statistical Analysis
One-way analysis of variance (ANOVA) with homogeneity of variances test was employed for the statistical analysis of baseline data characteristics. Given that the CRP detection threshold was set at 5, and values below 5 could not be quantified precisely, we designated all values below 5 as 2.5 for the purpose of quantitative statistical analysis. Taking into account the CRP testing standard established in our laboratory, where CRP levels greater than 5 are considered elevated compared to the normal population,1,13 we proceeded to classify individuals with CRP levels above 5 as elevated (positive) and those with levels below 5 as normal (negative). Subsequently, we conducted qualitative statistical analyses once again. We calculated diagnostic performance metrics including sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV), along with their corresponding 95% confidence intervals. We then employed the McNemar method to assess statistical differences. A p-value less than 0.05 was defined as indicating statistical significance. All statistical analyses were conducted using IBM SPSS version 26.0.
Results
Characteristics of Study Participants
A total of 78 patients with suspected spinal infection were included in the study based on their clinical symptoms, laboratory tests, and imaging.1,13 Ultimately, a total of 50 patients were categorized as infected cases based on the histopathological examination of the samples, while the remaining 28 patients were considered aseptic cases. No statistically significant differences were observed in terms of age and gender between the infection group and the aseptic group. The infection group showed significantly elevated levels of ESR, CRP, and WBC (P<0.001) compared to the aseptic group (Table 1).
 |
Table 1 Baseline Characteristics of Study participants |
Although no statistically significant difference was observed in age and gender between the two groups, the average age exceeded 54 years in both groups. In the present study, all identified pathogens were shown in Figure 1 and the most frequently identified pathogens were Mycobacterium tuberculosis (MTB), Staphylococcus aureus, and Escherichia coli. Compared to traditional culture-based diagnostic methods, mNGS detected a greater variety of bacteria and fungi, and only mNGS was capable of detecting the presence of viruses.
 |
Figure 1 Pathogens detected by mNGS and culture. |
The Diagnostic Performance of mNGS Compared to Culture in Spinal Infections
In the 28 aseptic samples, only one case yielded a positive culture result, identifying Staphylococcus epidermidis. This represents the sole case of a positive blood culture among our patients; all other positive culture results were derived from cultures of lesion samples. Simultaneously, mNGS identified Coxiella burnetii from one aseptic sample. In the remaining 26 aseptic samples, the detection results of mNGS and culture were both sterile, with complete consistency between the two methods.
Among the 50 infected samples, 16 yielded positive culture results, while 42 exhibited positive findings through mNGS (Figure 2). Among the 16 culture-positive cases, a total of 2 cases showed negative results by mNGS, with the culture results indicating Staphylococcus epidermidis and Escherichia coli, respectively. In the remaining 14 culture-positive cases, there were 2 cases where the culture results were completely discordant with the mNGS findings. The first case yielded Staphylococcus epidermidis in culture, while mNGS detected only Aspergillus niger. In another case, Klebsiella pneumoniae was cultured, whereas mNGS exclusively identified Pseudomonas aeruginosa. Among the 12 cases with concordant results between culture and mNGS, 8 cases showed complete agreement, while 4 cases exhibited partial concordance. In the 4 cases with partially concordant results, culture only captured a single bacterium present in the mNGS detection results.
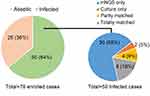 |
Figure 2 Comparison of diagnostic efficacy between mNGS and culture in spinal infection patients. |
Overall, in the diagnosis of spinal infections, mNGS demonstrated significantly higher sensitivity and NPV compared to culture, while the specificity and PPV of the two methods did not exhibit significant differences (Table 2).
 |
Table 2 Comparison of mNGS and Conventional Culture in Spinal Infection |
The Diagnostic Performance of mNGS Compared to Culture in Spinal TB
In the present study, the most commonly pathogen was MTB. Therefore, we conducted further analysis focused on this bacterium. Among the 11 spinal TB patients, both mNGS and T-spot detected 10 cases, while culture did not yield any positive results. In the diagnosis of spinal TB, mNGS showed significantly higher sensitivity compared to culture, while the specificity, PPV and NPV of the two methods did not exhibit significant differences (Table 3). In addition, despite the similar sensitivity between T-spot and mNGS, the specificity of mNGS was significantly higher than that of T-spot (Table 3).
 |
Table 3 Comparison of mNGS, T-Spot and Conventional Culture in Spinal TB |
Discussion
With the gradual popularization of mNGS technology and the reduction in its testing costs, it is increasingly being applied to clinical diagnostics. Due to the diagnostic performance of mNGS in spinal infection is still under investigation, the present study focuses on revealing its application value in the diagnosis of spinal infections. Firstly, among all confirmed pathogens, mNGS detected over 71% of the pathogens, while culture only detected approximately 14%. Secondly, among the 50 spinal infection patients, mNGS confirmed the presence of pathogens from 28 culture-negative cases. Moreover, in 4 culture-positive cases, mNGS additionally confirmed the presence of other pathogens. Thirdly, although the number of positive culture cases was limited, among the 16 cases with positive culture results, mNGS showed concordance with the culture results in 14 cases. These advantages of mNGS in diagnosing spinal infections are consistent with previous relevant research.10,14,15
Laboratory indicators such as ESR, WBC, and CRP presents notable elevations in the spinal infection group. The elevation of these indicators is helpful in raising suspicion of spinal infection, aligning with the guidance provided by the Infectious Diseases Society of America (IDSA) for infection diagnosis and treatment.13 However, the definitive diagnosis and treatment of spinal infections rely on methods such as culture to confirm the specific pathogens. The low sensitivity of culture and the difficulty in obtaining spinal infection lesion samples both contribute to the challenging nature of diagnosis.16,17 In addition, patients with suspicion of infection often arrive at our hospital after having empirically used antibiotics at county or lower-level hospitals, further reducing the success rate of culture. The low sensitivity of culture observed in this study can be explained, and it is consistent with similar studies.18,19 Thus, clinicians should adhere to the IDSA guideline (2015), reserving antibiotic use until pathogen identification except in cases of urgent antibiotic and surgical treatment. However, as reported in other literature, mNGS maintains high sensitivity even in the presence of antibiotic interference.20,21 Given the reality of antibiotic abuse and the challenges in obtaining spinal lesion samples, the results of the present study support the use of mNGS as a significant complementary diagnostic approach to culture.
The present study identified MTB and Staphylococcus aureus as the most common pathogens, a result similar to previously published research from China.10,15 In the present study, culture did not confirm any MTB, while mNGS confirmed 10 out of 11 in spinal TB patients. The reason for such a low success rate in culture could be attributed to the fact that our hospital laboratory concludes culturing after 7 days, while culturing for tuberculosis often requires more than 10 days.22 Utilizing egg-based or BACTEC 460 TB system could potentially shorten the culture time to around 9 days.22 To obtain positive culture results, you could either utilize specialized culture media for MTB or extend the culture duration appropriately. However, mNGS could provide test results within 48 hours of sample acquisition, enabling more efficient early-stage diagnosis compared to culture. Additionally, T-spot exhibited better sensitivity and shorter detection time in tuberculosis testing compared to culture, making it an important auxiliary diagnostic tool for MTB. According to the Table 3, although T-spot and mNGS demonstrated similar sensitivity and comparable detection times in MTB testing, mNGS held an advantage in terms of specificity. Therefore, the results of the present study supported focusing on both diagnostic approaches when suspecting spinal TB.
For the clinical practice of spinal infection diagnosis and treatment, early diagnosis was of paramount importance. Early identification of the causative pathogen could reduce patient suffering, minimize the occurrence of deformities, and even lead to a complete cure with conservative treatment. Though the IDSA guideline (2015) had mentioned some promising technology for spinal infection such as mNGS, there was still lack of clinical evidence to confirm its clinical efficacy. The results of this study robustly confirmed the diagnostic efficacy of mNGS in spinal infections from three aspects: time, accuracy, and comprehensiveness.
However, in this study, one case in the aseptic group showed positive culture results and another showed positive mNGS results. Firstly, one case in the aseptic group cultured Staphylococcus epidermidis, and this patient was ultimately diagnosed with malignant tumor. The positive culture result for this patient was obtained from blood culture upon admission. The patient presented with symptoms of fever and a cold at the time of admission. However, considering the specificity of Staphylococcus epidermidis (commonly associated with specimen contamination), this result may be related to the patient’s cold and fever symptoms or could potentially be attributed to specimen contamination. The other aseptic patient was determined by mNGS to have Coxiella burnetii, and this patient was also ultimately diagnosed with a malignant tumor. Although the primary focus of the specimen in this patient was the tumor, there might have been a low concentration of bacterial colonies present, which did not form a clear infectious lesion confirmed by pathology and culture.
Throughout this study, there were instances of discrepancies between culture and mNGS results. We identified two cases (Figure 2) where the culture yielded positive results while mNGS indicated negative findings, encompassing various bacterial species (Figure 1). Upon scrutinizing our procedural workflow and relevant literature on mNGS, we pinpointed several potential influencing factors: 1) The cost disparity between culture and mNGS, where culture expenses are only 1/60th of mNGS. As a result, multiple samples were cultured, while mNGS utilized a single sample from the lesion alone. The diversity among these different samples likely contributed to the reduced success rate of mNGS. 2) Nucleic acid degradation in the submitted samples and potential interference from background bacteria might have influenced the diagnostic outcomes of mNGS.23,24
The present study has certain strengths and limitations. Firstly, to our knowledge, recent studies regarding the application of mNGS in spinal infection have been limited by small sample sizes.10,25 Despite the relatively recent adoption of mNGS technology in clinical practice and the relatively low prevalence of spinal infection cases compared to other types of infections, the present study managed to include a total of 78 cases, thereby addressing potential issues related to sample size insufficiency. The second strength lies in the fact that the mNGS can be performed in-hospital, reducing transportation time, mitigating nucleic acid degradation and minimizing contamination risks. This has resulted in less susceptibility of mNGS to external factors. However, there are still certain limitations in the study that should be noted. Due to the lack of adequate hardware for implementing mNGS in primary healthcare facilities, multi-center collaboration might yield varying research outcomes. Additionally, because this study was retrospective, some patients with suspected spinal infections were not included in the study because they did not complete mNGS for financial or other reasons, which may have resulted in some study bias.
Conclusion
Herein, this study demonstrates the advantages of mNGS technology in terms of accuracy and comprehensiveness in diagnosing spinal infections. Beyond its advantages in bacterial detection, mNGS exhibits a distinct advantage in the detection of fungi and viruses. Using mNGS as an additional diagnostic method in spinal infection diagnosis will be benefit for patients. Besides, this study provides additional clinical evidence for the application of mNGS in clinical practice.
Data Sharing Statement
If necessary, data can be obtained from the corresponding author upon resonable request.
Ethical Approval and Consent to Participate
All procedures were performed in compliance with relevant laws and institutional guidelines and that the medical ethics committee of Sun Yat-Sen Memorial Hospital of Sun Yat-sen University has approved them (SYSKY-2023-1095-01). At the time of admission for each patient, we have obtained informed consent from all patients. Authors confirmed that the present study strictly adheres to the relevant provisions of the Declaration of Helsinki.
Funding
Professor Li has received funding from Guangzhou Science and technology projects (Grant numbers: 202102010259). Doctor Liu has received funding from Jointly Sponsored Project by Municipal and University (Institute) Foundation for Basic and Applied Basic Research (Grant numbers: 202201020531).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Duarte RM, Vaccaro AR. Spinal infection: state of the art and management algorithm. Eur Spine J. 2013;22(12):2787–2799. doi:10.1007/s00586-013-2850-1
2. Gouliouris T, Aliyu SH, Brown NM. Spondylodiscitis: update on diagnosis and management. J Antimicrob Chemother. 2010;65(Suppl 3):iii11–iii24. doi:10.1093/jac/dkq303
3. Lew DP, Waldvogel FA. Osteomyelitis. Lancet. 2004;364(9431):369–379. doi:10.1016/S0140-6736(04)16727-5
4. Lener S, Hartmann S, Barbagallo GMV, Certo F, Thome C, Tschugg A. Management of spinal infection: a review of the literature. Acta Neurochir. 2018;160(3):487–496. doi:10.1007/s00701-018-3467-2
5. Han D, Li Z, Li R, Tan P, Zhang R, Li J. mNGS in clinical microbiology laboratories: on the road to maturity. Crit Rev Microbiol. 2019;45(5–6):668–685. doi:10.1080/1040841X.2019.1681933
6. Sun L, Zhang S, Yang Z, et al. Clinical application and influencing factor analysis of Metagenomic Next-Generation Sequencing (mNGS) in ICU patients with sepsis. Front Cell Infect Microbiol. 2022;12:905132. doi:10.3389/fcimb.2022.905132
7. Cheng H, Tan N, Lu Y, et al. Lumbar spine infection with eikenella corrodens presented as abdominal pain: a case report and literature review. Infect Drug Resist. 2023;16:1407–1417. doi:10.2147/IDR.S400451
8. Yu C, Guo W, Zhang Z, et al. The impact of mNGS technology in the etiological diagnosis of severe pneumonia in children during the epidemic of COVID-19. Infect Drug Resist. 2023;16:2395–2402. doi:10.2147/IDR.S403851
9. Gokdemir FS, Iseri OD, Sharma A, Achar PN, Eyidogan F. Metagenomics Next Generation Sequencing (mNGS): an exciting tool for early and accurate diagnostic of fungal pathogens in plants. J Fungi. 2022;8(11):1195. doi:10.3390/jof8111195
10. Ma C, Wu H, Chen G, Liang C, Wu L, Xiao Y. The potential of metagenomic next-generation sequencing in diagnosis of spinal infection: a retrospective study. Eur Spine J. 2022;31(2):442–447. doi:10.1007/s00586-021-07026-5
11. Yang X, Wu W, Wang Y, Wu W, Huang X, Xu L. A case of secondary pulmonary syphilis - the utility of mNGS in bronchoalveolar lavage fluid: a case report. Infect Drug Resist. 2022;15:5215–5219. doi:10.2147/IDR.S373711
12. Miao Q, Ma Y, Wang Q, et al. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis. 2018;67(suppl_2):S231–S240. doi:10.1093/cid/ciy693
13. Berbari EF, Kanj SS, Kowalski TJ, et al. 2015 Infectious Diseases Society of America (IDSA) clinical practice guidelines for the diagnosis and treatment of native vertebral osteomyelitis in adults. Clin Infect Dis. 2015;61(6):e26–e46. doi:10.1093/cid/civ482
14. Li Y, Yao XW, Tang L, et al. Diagnostic efficiency of metagenomic next-generation sequencing for suspected spinal tuberculosis in China: a multicenter prospective study. Front Microbiol. 2022;13:1018938. doi:10.3389/fmicb.2022.1018938
15. Wang C, Hu J, Gu Y, Wang X, Chen Y, Yuan W. Application of next-generation metagenomic sequencing in the diagnosis and treatment of acute spinal infections. Heliyon. 2023;9(3):e13951.
16. Tsantes AG, Papadopoulos DV, Vrioni G, et al. Spinal infections: an update. Microorganisms. 2020;8(4):476. doi:10.3390/microorganisms8040476
17. Urrutia J, Bono CM. Update on the diagnosis and management of spinal infections. Instr Course Lect. 2022;71:439–449.
18. Wan J, Duan L, Chen Q, et al. Potential clinical impact of metagenomic next-generation sequencing of plasma for cervical spine injury with sepsis in intensive care unit: a retrospective study. Front Cell Infect Microbiol. 2022;12:948602. doi:10.3389/fcimb.2022.948602
19. Xu L, Zhou Z, Wang Y, Song C, Tan H. Improved accuracy of etiological diagnosis of spinal infection by metagenomic next-generation sequencing. Front Cell Infect Microbiol. 2022;12:929701. doi:10.3389/fcimb.2022.929701
20. Wang G, Long J, Zhuang Y, et al. Application of metagenomic next-generation sequencing in the detection of pathogens in spinal infections. Spine J. 2023;23(6):859–867. doi:10.1016/j.spinee.2023.02.001
21. Zheng L, Kang Z, Wang R, et al. Evaluation of the diagnostic performance of mNGS in detecting intra-abdominal infections of the emergency department patients. Infect Drug Resist. 2023;16:1421–1432. doi:10.2147/IDR.S396699
22. Leowattana W, Leowattana P, Leowattana T. Tuberculosis of the spine. World J Orthop. 2023;14(5):275–293. doi:10.5312/wjo.v14.i5.275
23. Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4(4):663–674. doi:10.1038/s41564-018-0349-6
24. Miller S, Naccache SN, Samayoa E, et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 2019;29(5):831–842. doi:10.1101/gr.238170.118
25. Zhang Y, Chen J, Yi X, et al. Evaluation of the metagenomic next-generation sequencing performance in pathogenic detection in patients with spinal infection. Front Cell Infect Microbiol. 2022;12:967584. doi:10.3389/fcimb.2022.967584
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
