Back to Journals » Journal of Hepatocellular Carcinoma » Volume 11
Development of a Reliable GADSAH Model for Differentiating AFP-negative Hepatic Benign and Malignant Occupying Lesions
Authors Long X, Zeng H, Zhang Y, Lu Q, Cao Z, Shu H
Received 8 December 2023
Accepted for publication 9 March 2024
Published 23 March 2024 Volume 2024:11 Pages 607—618
DOI https://doi.org/10.2147/JHC.S452628
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr David Gerber
Xiaoling Long,1,* Huan Zeng,1,* Yun Zhang,1 Qiulong Lu,1 Zhao Cao,2 Hong Shu1
1Department of Clinical Laboratory, Guangxi Medical University Cancer Hospital, Nanning, Guangxi, 530021, People’s Republic of China; 2Department of Clinical Laboratory, First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi, 530021, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hong Shu, Department of Clinical Laboratory, Guangxi Medical University Cancer Hospital, No. 71 Hedi Road, Qingxiu District, Nanning, Guangxi, 530021, People’s Republic of China, Tel +86 7715640296, Email [email protected] Zhao Cao, Department of Clinical Laboratory, First Affiliated Hospital of Guangxi Medical University, No. 6 Shuangyong Road, Qingxiu District, Nanning, Guangxi, 530021, People’s Republic of China, Tel +86 7715356724, Email [email protected]
Purpose: Developing a high-value, convenient, and validated differential diagnosis model to differentiate alpha-fetoprotein (AFP) negative hepatic occupying lesions and assist clinicians in early identification and intervention.
Patients and Methods: A total of 340 patients with AFP-negative hepatic occupying lesions who were admitted to the Guangxi Medical University Cancer Hospital between August 2021 and April 2023 were included in the final retrospective analysis. The data were randomly divided into training and validation sets in a 7:3 ratio after performing multiple interpolations. In the training set, laboratory variables and models were screened using least absolute shrinkage and selection operator regression analysis, comparison of five machine learning algorithms, and univariate, as well as multivariate logistic regression analysis. A diagnostic prediction nomogram model was developed. We evaluated and validated the model using the receiver operating characteristic (ROC) curve analysis, calibration curve analysis, and decision curve analysis (DCA).
Results: We identified six significant predictive factors from the results of multivariate logistic analysis in the training set and incorporated them into the nomogram model for diagnosing AFP-negative hepatic malignant occupying lesions (HMOL). The diagnostic nomogram, including gender, age, des-gamma-carboxy prothrombin (DCP), serum ferritin (SF), AFP, and hepatitis B surface antigen (HBsAg), achieved an area under the curve of 0.905 discriminated patients with HMOL from those with benign occupying lesions. Additionally, calibration curves demonstrated the close alignment between the nomogram predictions and the ideal curve, along with the consistency between predictions and actual results. Moreover, the DCA curves illustrated indicated benefit for all patients. These finding were confirmed by the validation set.
Conclusion: The GADSAH model specifically targets the discrimination of malignant and benign liver lesions in AFP-negative patients. It offers a noninvasive, cost-effective, and efficient approach for diagnosing such cases.
Keywords: hepatic occupying lesions, alpha-fetoprotein-negative, nomogram, diagnostic model
Introduction
Hepatic space-occupying lesions are abnormal tissue areas, comprising solid or cystic masses, located with the liver. Depending on the characteristics of the lesion, it can be categorized as either a malignant or a benign hepatic lesion.1 However, it is essential to note that the treatment and prognosis for these two types of lesions differ significantly. Hepatic malignant occupying lesions (HMOL), primarily hepatocellular carcinoma (HCC), ranked sixth in incidence and third in mortality among global cancer diseases, as reported in the 2020 Global Cancer Statistics.2,3 The incidence and mortality rates of HCC have shown a consistent increase worldwide over the last two decades.4 Late diagnosis often leads to a poor prognosis for patients. Timely detection allows for liver transplant or surgical resection, which can result in a 5-year survival rate exceeding 70%.5 In contrast, hepatic benign occupying lesions (HBOL), such as hepatic hemangiomas, generally exhibit slow growth and often require mere observation or local intervention.6 Therefore, an accurate differential diagnosis of hepatic occupying lesions is crucial for developing appropriate treatment plans and predicting prognosis.
The Current guidelines recommend screening for HCC using alpha-fetoprotein (AFP) and B-ultrasound.7 However, as the proportion of small hepatocellular carcinomas being diagnosed continues to increase, the sensitivity of AFP progressively decreases.8,9 Furthermore, studies have shown that approximately 30% to 40% of HCC cases are AFP-negative.10 Additionally, ultrasound has limited sensitivity for assessing HMOL.11,12 Other imaging modalities, especially in the detection of small HCC and differentiation from other benign lesions, still lead to misdiagnosis and missed diagnoses.13,14 While histopathological diagnosis remains the gold standard for the diagnosing HMOL, it is an invasive and risky. In contrast, blood diagnostic markers offer a non-invasive, low-cost, objective, and convenient approach to the screening and monitoring liver malignancies. Although new HMOL blood diagnostic markers have been developed, they are still in the research stage and have not been widely implemented in clinical practice.15–17 Furthermore, the combination of multiple blood markers and demographic characteristics to construct hepatocellular carcinoma prediction models and enhance the early diagnosis rate of HCC has been a popular research topic in recent years.18–20
Therefore, it is crucial to develop a rapid, convenient, and sensitive screening and surveillance method using a series of clinical examinations, in order to ensure timely detection of malignant lesions in asymptomatic individuals at risk. Machine learning, an emerging medical field of artificial intelligence in medicine, possesses the ability to process large and complex data, and thus can facilitate more precise disease diagnoses and personalize patient treatments.21 In the realm of personalized cancer therapy, nomograms serve as statistical tools that provide patients with a personalized prediction based on a set of variables. Nomogram models have been used to diagnose or predict cancer by integrating multiple parameters.22,23 However, little research has focused on developing a nomogram prediction model for differentiating between HMOL and HBOL for patients with AFP-negative. Therefore, we conducted a retrospective analysis of the clinical data and laboratory parameters from 245 cases of HMOL and 95 cases of HBOL patients admitted to the Guangxi Medical University Cancer Hospital. The objective was to construct a reliable nomogram model, which aims to distinguish AFP-negative HMOL from HBOL by using machine learning algorithms.
Materials and Methods
Patients
This study conducted a retrospective analysis of data from 1647 cases of hepatic occupying lesions collected at the Guangxi Medical University Cancer Hospital between August 2021 and April 2023. Inclusion criteria required patients to have received their initial diagnosis through imaging or pathology prior to surgical treatment.24,25 Exclusion criteria were as follows: 1. Received any anti-cancer treatment; 2. Co-existence of malignant localization in other organs; 3. Co-existence of benign and malignant occupations; 4. Diagnostic inconsistencies in pathology and imaging; 5. AFP≥20mg/mL cases. Ultimately, 340 cases of hepatic lesions were included in the study, consisting of 245 cases of HMOL and 95 cases of HBOL.
Clinicopathologic Characteristics
General patient data, routine hematological, biochemical and immunological parameters were collected from 340 individuals with hepatic occupying lesions. The collected information included gender, age, blood platelet (PLT), prothrombin time (PT), activated partial thromboplastin time (APTT), thrombin time (TT), fibrinogen (FIB), des-gamma-carboxy prothrombin (DCP), total bilirubin (TBIL), direct bilirubin (DBIL), indirect bilirubin (IBIL), serum ferritin (SF), alanine aminotransferase (ALT), aspartate transaminase (AST), alkaline phosphatase (ALP), lactate dehydrogenase (LDH), gamma-glutamyltransferase (γ-GGT), alpha-L-fucosidase (AFU), 5’-nucleotidase (5’-NT), monoamine oxidase (MAO), total bile acid (TBA), thymidine kinase 1 (TK1), heat-shock protein 90 alpha (HSP90α), complement protein 3 (C3), complement protein 4 (C4); carcinoembryonic antigen (CEA), carbohydrate antigen 125 (CA125), carbohydrate antigen 153 (CA153), carbohydrate antigen 199 (CA199), AFP, lens culinaris agglutinin-reactive fraction of AFP (AFP-L3), lens culinaris agglutinin-reactive fraction of AFP/total AFP (AFP-L3%), hepatitis B surface antigen (HBsAg), and hepatitis B e antigen (HBeAg).
Statistical Analysis
Data Preprocessing
All calculations were performed using R software (version 4.3.1) and relevant packages. Missing data points below 20% were imputed using the predictive mean matching method.26
Model Building
Significant variables in the analysis of differences between the HBOL cohort and the HMOL cohort were screened. The two cohorts were randomly divided into training and validation sets using a 7:3 ratio. In the training set, the least absolute shrinkage and selection operator (LASSO) were used to screen variables.27 To screen the best predictive modeling methods, five common machine learning algorithms were selected, including decision tree (DT), logistic regression (LR), neural network (NNET), random forest (RF), and support vector machine (SVM) to evaluate the performance of the model in the training set.28 The performance of models for the statistically significant features was assessed using area under the curve receiver operating characteristics curve (AUROC). Therefore, univariate and bidirectional elimination logistic regression analyses were conducted on chosen the significant factors to assess the final model. Finally, a predictive nomogram was constructed to discriminate AFP-negative HMOL and HBOL. The LASSO regression analysis applied ten-fold cross-validation, and five machine learning algorithms used ten-fold bootstrap validation.
Model Evaluation
The performance of the nomograms was assessed using AUROC and calibration curve analyses. The calibration capability of the model was evaluated using the Hosmer-Lemeshow test, and the calibration curves were analyzed using 1000 bootstraps. Additionally, decision curve analysis (DCA) was performed to determine the predicted net benefit threshold. These methods for evaluating models were also evaluated by the validation set.
Non-normally distributed data were presented as median and interquartile range. The chi-squared test was used for categorical variables, and the Mann–Whitney U-test was used for numerical variables. Results with a p value of less than 0.05 were considered significant.
Results
Patient Cohorts and Clinicopathologic Features
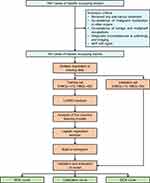
A total of 1647 cases of liver lesions were subjected to inclusion and exclusion criteria, resulting in a final cohort of 340 cases. The demographics and clinicopathologic characteristics of the 340 patients were listed in Table S1. Among these patients, 95 (27.94%) cases were diagnosed as benign and 245 (72.06%) were diagnosed as malignant. To address missing data, multiple imputation methods were utilized as a preprocessing step. Subsequently, statistical significance tests were performed on all clinical characteristic parameters. The detailed process is presented in a flow diagram (Figure 1). Within the AFP-negative HBOL and HMOL cohorts, there were 30 parameters that exhibited statistically significant variances among a total of 35 parameters (P<0.05, Table 1). Significantly, there is a notable difference in the distribution of benign and malignant liver lesions between genders (P<0.001, Table 1). In each cohort, the majority of patients with HMOL were male (85.31% vs 14.69%), while the majority of patients with HBOL were female (58.95% vs 41.05%), and the median age surpassed 45 years (48 vs 55). Furthermore, several parameters including Age, PT, TT, FIB, D-Dimer, log10(DCP), TBIL, DBIL, SF, ALT, AST, ALP, LDH, γ-GGT, AFU, 5-NT, TBA, TK1, HSP90α, C4, CEA, CA125, CA153, CA199, log10(AFP), AFP-L3, HBsAg, and HBeAg were found to be higher in the HMOL cohort compared to the HBOL cohort (P<0.05, Table 1). Conversely, PLT level was lower in the HMOL cohort (P<0.05, Table 1). The patients were divided into two sets, namely the training set and the validation set, comprising 237 and 103 patients, respectively. Importantly, all thirty variables mentioned above were found to be statistically significant. Notably, there were no significant differences were observed between the parameters in these two sets (P≥0.05, Table S2).
 |
Table 1 Comparison of Variables Between HBOL Cohort and HMOL Cohort |
Clinical Parameters and Method Selection for the Predictive Model
In the training set, the LASSO regression analysis was performed on 30 variables (Figure 2A and B). The dashed line in Figure 2B represents the maximum value for lambda (λ) within one standard error of the minimum binomial deviation. At λ = 0.061, six variables with non-zero coefficients were identified via ten-fold cross-validated LASSO regression analysis as concise combinations distinguishing AFP-negative HBOL from HMOL. The variables contained gender, age, log10(DCP), SF, log10(AFP), and HBsAg, as presented in Table S3. Subsequently, five machine learning models were analyzed by the above six variables. The receiver operating characteristic (ROC) curve of the training set demonstrated that the area under the curve (AUC) value of DT, LR, NNET, RF, and SVM models in diagnosing AFP-negative HMOL were 0.790, 0.898, 0.775, 0.896, and 0.888, respectively (Figure 2C). Notably, the LR model displayed the most significant diagnostic ability, with an AUC of 0.898 for distinguishing between benign and malignant hepatic lesions.
Logistic Regression Analysis and Model Building
Significance variables identified in univariate logistic regression analysis of the training set included gender, age, log10(DCP), SF, log10(AFP), and HBsAg (P<0.05, Table 2), and were subsequently subjected to the multivariate regression analysis. Bidirectional elimination regression analysis revealed that the final model with the smallest Akaike information criterion (AIC), included gender (odds ratio [OR] = 2.85, 95% confidence interval [CI] 1.24–6.57), age (OR = 1.03, 95% CI 0.99–1.07), log10(DCP) (OR = 2.97, 95% CI 1.67–5.28), SF (OR = 1.01, 95% CI 1.01–1.01), log10(AFP) (OR = 4.46, 95% CI 0.74–27.03) and HBsAg (OR = 1.01, 95% CI 1.01–1.01), as shown in Table 2. The AUROCs of gender (AUC=0.727), age (AUC=0.641), log10(DCP) (AUC=0.799), SF (AUC=0.801), log10(AFP) (AUC=0.741) and HBsAg (AUC=0.758) were all above 0.6 (Table S4). The accuracy, sensitivity, and specificity of these parameters can be found in Table S4. Based on these six independent predictors, a LR equation for AFP-negative HMOL diagnosis was established as follows: 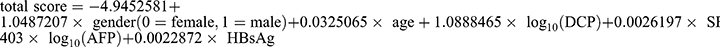 . The nomogram provided an example of the correctly predicting an AFP-negative HMOL diagnostic result with an optimal cut-off value of 0.62436 (Figure 3, Table S4).
. The nomogram provided an example of the correctly predicting an AFP-negative HMOL diagnostic result with an optimal cut-off value of 0.62436 (Figure 3, Table S4).
 |
Table 2 Results of Logistic Regression Analysis |
Validation of the Model and Evaluation of Clinical Application
The AUC value of the nomogram model in the training set is 0.905, with a sensitivity is 86.6%, a specificity is 80.0% and an accuracy is 84.8%. In the validation set, the AUC value, sensitivity, specificity and accuracy of the nomogram model are 0.880, 82.2%, 86.7%, and 83.5% respectively (Figure 4A and B, Table S4). The GADSAH, serving as a discrimination model, outperformed any single variable model in accurately differentiating various types of focal liver lesions and providing valuable insights for clinical decision-making (Table S4). To further validate the prediction results, we conducted internal validation using the bootstrap approach. The Hosmer-Lemeshow test for the final model demonstrated good fit in both the training and validation sets, indicating a well-fitted model (P>0.05, Figure 4C and D). Moreover, the prediction and calibration curves of the nomogram model closely resembled the ideal curves, confirming its strong predictive ability and consistency (Figure 4C and D). Additionally, decision curve analyses revealed that the patient benefit curve for our model surpassed the two extreme curves (grey and black) under the same risk threshold probability region, suggesting a greater net benefit and clinical utility of the model (Figure 4E and F).
Discussion
In this study, we developed a nomogram model called GADSAH to differentiate between AFP-negative HBOL and HMOL. This model integrates six key predictive factors, encompassing a range of demographic and laboratory parameters including gender, age, log10(DCP), SF, log10(AFP), and HBsAg. Our nomogram model accurately predicted the diagnosis of AFP-negative HMOL with an optimal cut-off value. The calibration curve closely resembled the ideal standard curve, revealing that the nomogram model possessed adequate statistical capability to distinguish between benign and malignant hepatic occupying lesions.
Furthermore, our GADSAH model exhibited excellent performance in diagnosing AFP-negative HMOL, demonstrating heightened sensitivity and specificity. Misdiagnosing some HBOL cases as hepatocellular carcinoma can lead to unnecessary treatment, financial burden, and diminished quality of life for patients.14,25 Additionally, negative results from AFP tests can complicate the distinction between HMOL and HBOL. Thus, our model can assist surgeons in designing tailored treatment plans prior to initiating treatment. By employing prospective quantitative prediction, the precision of diagnosing AFP-negative HMOL can be enhanced while simultaneously preventing overtreatment of HBOL.
According to relevant studies using world-standard population calculations, the incidence of primary liver cancer in males is 3.28%, surpassing the 1.42% incidence in females.3 Consequently, the incidence of liver cancer is higher in males than in females.29 However, HBOL is less prevalent in men compared to women.30 This difference between gender can be attributed to various risk factors associated with liver cancer, including increased susceptibility to hepatitis B and C viruses, history of smoking and alcohol consumption, higher levels of iron stores, and elevated blood levels of aflatoxin and androgens.31 Consistent with this, our study revealed that men have a higher risk of developing HMOL than women.
As a storage form of iron in the body,32 there is increasing evidence that SF is significantly increased in patients with liver cancer, and serves an important marker for its diagnosis, particularly in patients with AFP negative liver cancer.33–35 In line with this, incorporating SF into the model allows for the detection of AFP-negative HMOL among HBOL.
AFP is commonly used as a tumor marker for HCC screening and clinical diagnosis.36 However, its accuracy as a serological marker for detecting liver malignant tumors is limited.11,37,38 Previous studies have primarily focused on distinguishing HCC from patients with Hepatitis B-related benign liver disease. Due to the large number of occupying lesions patients admitted to our cancer hospital, the differentiation between AFP-negative HMOL and HBOL is worthy of great attention. Therefore, we excluded cases that were positive for AFP and concentrated on differentiating between AFP-negative HMOL from HBOL. The Food and Drug Administration (FDA) in the United States has approved DCP as a diagnostic marker for liver cancer in East Asian countries. DCP has high specificity in the diagnosis of liver cancer, which is useful in distinguishing between benign and malignant tumors. The combined detection of DCP and AFP is more sensitive than single detection in early HCC detection.39,40 Our study found that both AFP and DCP were independent risk factors for predicting AFP-negative HMOL, consistent with previous studies.
HBV infection is a significant risk factor for liver cancer development, accounting for more than two-thirds of cases in China.41 serum HBsAg detection plays a crucial role in HBV infection diagnosis.42 Numerous studies have reported the substantial diagnostic and predictive value of assessing serum HBsAg in liver malignancy.22,43,44 Our findings align with these established conclusions.
The nomogram model we developed, GADSAH depicts AFP combined with DCP as well as other readily available clinical parameters and predicts the probability of AFP-negative HMOL. Previous AFP combined with DCP multiparameter models have also been reported, such as the GALAD (gender, age, AFP-L3, AFP, and DCP) and ASAP (age, sex, AFP, and DCP) models have shown diagnostic value in identifying HCC within the context of chronic liver disease, particularly in cases with negative AFP results. The applicability of these models varies across different patient populations. Currently, there is a scarcity of reports establishing nomograms based on clinical parameters for distinguishing between benign and malignant liver lesions, especially for AFP-negative patients. While certain research studies have demonstrated that radiomics or ultrasound models can effectively discriminate between benign and malignant hepatic occupying lesions, the generalizability of such models in clinical practice is limited due to potential deficiencies in professionalism with new imaging techniques and the high economic burden on patients.45–47 Our GADSAH model focuses solely on discriminating between malignant and benign liver occupying lesions in AFP-negative patients, providing a noninvasive, cost-effective, and efficient method in diagnosing such cases. However, it is important to acknowledge the limitations of our study, which was a retrospective analysis conducted at a single center and lacks external validation. To address these limitations and further enhance the understanding of our model’s effectiveness and potential limitations, additional studies should be conducted in the future.
Conclusion
In conclusion, we have developed and validated a robust and objective nomogram model, known as GADSAH model which incorporates gender, age, and four routine clinical hematological parameters (log10(DCP), SF, log10(AFP), and HBsAg). This model effectively differentiates AFP-negative HMOL patients from the population with HBOL. The GADSAH model demonstrates excellent consistency, discrimination, and clinical relevance. It offers a rapid and accurate approach for identifying AFP-negative HMOL cases, thereby facilitating the implementation of targeted interventions and treatment protocols.
Abbreviations
HMOL, hepatic malignant occupying lesions; HCC, hepatocellular carcinoma; HBOL, hepatic benign occupying lesions; AFP, alpha-fetoprotein; PLT, blood platelet; PT, prothrombin time; APTT, activated partial thromboplastin time; TT, thrombin time; FIB, fibrinogen; DCP, des-gamma-carboxy prothrombin; TBIL, total bilirubin; DBIL, direct bilirubin; IBIL, indirect bilirubin; SF, serum ferritin; ALT, alanine aminotransferase; AST, aspartate transaminase; ALP, alkaline phosphatase; LDH, lactate dehydrogenase; γ-GGT, gamma-glutamyltransferase; AFU, alpha-L-fucosidase; 5’-NT, 5’-nucleotidase; MAO, monoamine oxidase; TBA, total bile acid; TK1, thymidine kinase 1; HSP90α, heat-shock protein 90 alpha; C3, Complement protein 3; C4, Complement protein 4; CEA, carcinoembryonic antigen; CA125, carbohydrate antigen 125; CA153, carbohydrate antigen 153; CA199, carbohydrate antigen 199; AFP-L3, lens culinaris agglutinin-reactive fraction of AFP; AFP-L3%, lens culinaris agglutinin-reactive fraction of AFP/total AFP; HBsAg, hepatitis B surface antigen; HBeAg, hepatitis B e antigen; LASSO, least absolute shrinkage and selection operator; DT, decision tree; LR, logistic regression; NNET, neural network; RF, random forest; SVM, support vector machines; AUROC, area under the receiver operating characteristic curve; DCA, decision curve analysis; ROC, receiver operating characteristic; AUC, area under the curve; OR, odds ratio; CI, confidence interval.
Data Sharing Statement
On reasonable request, the corresponding author will provide the datasets generated during and/or analyzed during the current study.
Ethics Statement
This study complied with the Declaration of Helsinki. Approval for this study was granted by the Medical Ethics Committee of Guangxi Medical University Cancer Hospital, and written informed consent was obtained from all participants. (Ethics approval number: LW2019043).
Consent to Publish
All authors contributed to the article and approved the submitted version.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Natural Science Fund Project of Guangxi Province of China (2018GXNSFAA281053, 2023GXNSFAA026038).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Marrero JA, Ahn J, Rajender Reddy K. ACG clinical guideline: the diagnosis and management of focal liver lesions. Am J Gastroenterol. 2014;109(9):1328–1347; quiz 1348. doi:10.1038/ajg.2014.213
2. Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nature Reviews Disease Primers. 2021;7(1):6. doi:10.1038/s41572-020-00240-3
3. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca a Cancer J Clinicians. 2021;71(3):209–249. doi:10.3322/caac.21660
4. Liu J, Tang W, Budhu A, et al. A viral exposure signature defines early onset of hepatocellular carcinoma. Cell. 2020;182(2):317–328.e310. doi:10.1016/j.cell.2020.05.038
5. Takayama T, Makuuchi M, Kojiro M, et al. Early hepatocellular carcinoma: pathology, imaging, and therapy. Ann Surg Oncol. 2008;15(4):972–978. doi:10.1245/s10434-007-9685-0
6. Luo L, Wang T, Cheng M, et al. Rare benign liver tumors that require differentiation from hepatocellular carcinoma: focus on diagnosis and treatment. J Cancer Res Clin Oncol. 2023;149(7):2843–2854. doi:10.1007/s00432-022-04169-w
7. Heimbach JK, Kulik LM, Finn RS, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67(1):358–380. doi:10.1002/hep.29086
8. Zhang G, Ha SA, Kim HK, et al. Combined analysis of AFP and HCCR-1 as an useful serological marker for small hepatocellular carcinoma: a prospective cohort study. Dis. Markers. 2012;32(4):265–271. doi:10.3233/dma-2011-0878
9. Farinati F, Marino D, De Giorgio M, et al. Diagnostic and prognostic role of alpha-fetoprotein in hepatocellular carcinoma: both or neither? Am J Gastroenterol. 2006;101(3):524–532. doi:10.1111/j.1572-0241.2006.00443.x
10. Chi X, Jiang L, Yuan Y, et al. A comparison of clinical pathologic characteristics between alpha-fetoprotein negative and positive hepatocellular carcinoma patients from Eastern and Southern China. BMC Gastroenterol. 2022;22(1):202. doi:10.1186/s12876-022-02279-w
11. Tzartzeva K, Obi J, Rich NE, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology. 2018;154(6):1706–1718.e1701. doi:10.1053/j.gastro.2018.01.064
12. Benhammou JN, Rich NE, Cholankeril G, et al. DETECT: Development of Technologies for Early HCC Detection. Gastroenterology. 2022;163(1):21–27 doi:10.1053/j.gastro.2022.03.024
13. Ariff B, Lloyd CR, Khan S, et al. Imaging of liver cancer. World J Gastroenterol. 2009;15(11):1289–1300. doi:10.3748/wjg.15.1289
14. Baranes L, Chiaradia M, Pigneur F, et al. Imaging benign hepatocellular tumors: atypical forms and diagnostic traps. Diagn. Interventional Imaging. 2013;94(7–8):677–695. doi:10.1016/j.diii.2013.05.002
15. Cao X, Cao Z, Ou C, et al. Combination of serum paraoxonase/arylesterase 1 and antithrombin-III is a promising non-invasion biomarker for discrimination of AFP-negative HCC versus liver cirrhosis patients. Clin Res Hepatol Gastroenterol. 2021;45(5):101583. doi:10.1016/j.clinre.2020.11.013
16. Shu H, Zhang L, Chen Y, et al. Quantification of Intact O-Glycopeptides on haptoglobin in sera of patients with hepatocellular carcinoma and liver cirrhosis. Front Chem. 2021;9:705341. doi:10.3389/fchem.2021.705341
17. Kim DY, Toan BN, Tan CK, et al. Utility of combining PIVKA-II and AFP in the surveillance and monitoring of hepatocellular carcinoma in the Asia-Pacific region. Clin mol hepatol. 2023;29(2):277–292. doi:10.3350/cmh.2022.0212
18. Fan R, Papatheodoridis G, Sun J, et al. aMAP risk score predicts hepatocellular carcinoma development in patients with chronic hepatitis. J Hepatol. 2020;73(6):1368–1378. doi:10.1016/j.jhep.2020.07.025
19. Yang T, Xing H, Wang G, et al. A novel online calculator based on serum biomarkers to detect hepatocellular carcinoma among patients with hepatitis B. Clin. Chem. 2019;65(12):1543–1553. doi:10.1373/clinchem.2019.308965
20. Luo QQ, Li QN, Cai D, et al. The Index sAGP is valuable for distinguishing atypical hepatocellular carcinoma from atypical benign focal hepatic lesions. J Hepatocell Carcinoma. 2024;11:317–325. doi:10.2147/jhc.S443273
21. Handelman GS, Kok HK, Chandra RV, Razavi AH, Lee MJ, Asadi H. eDoctor: machine learning and the future of medicine. J Intern Med. 2018;284(6):603–619. doi:10.1111/joim.12822
22. Liu ZJ, Xu Y, Wang WX, et al. Development and application of hepatocellular carcinoma risk prediction model based on clinical characteristics and liver related indexes. World j Gastroint Oncol. 2023;15(8):1486–1496. doi:10.4251/wjgo.v15.i8.1486
23. An S, Zhan X, Liu M, Li L, Wu J. Diagnostic and prognostic nomograms for hepatocellular carcinoma based on PIVKA-II and serum biomarkers. Diagnostics. 2023;13(8). doi:10.3390/diagnostics13081442
24. Bureau of Medical Administration. 原发性肝癌诊疗指南(2022年版) [Standardization for diagnosis and treatment of hepatocellular carcinoma (2022 edition)]. Zhonghua Gan Zang Bing Za Zhi. 2022;30(4):367–388. Chinese. doi:10.3760/cma.j.cn501113-20220413-00193
25. European Association for the Study of the Liver. EASL Clinical Practice Guidelines on the management of benign liver tumours. J Hepatol. 2016;65(2):386–398. doi:10.1016/j.jhep.2016.04.001
26. Austin PC, White IR, Lee DS, van Buuren S. Missing data in clinical research: a tutorial on multiple imputation. Can J Cardiol. 2021;37(9):1322–1331. doi:10.1016/j.cjca.2020.11.010
27. Liu Y, Han Y, Chen B, et al. A new online dynamic nomogram: construction and validation of an assistant decision-making model for laryngeal squamous cell carcinoma. Front Oncol. 2022;12:829761. doi:10.3389/fonc.2022.829761
28. Li M, Wang Q, Lu P, et al. Development of a machine learning-based prediction model for chemotherapy-induced myelosuppression in children with wilms’ tumor. Cancers. 2023;15(4). doi:10.3390/cancers15041078
29. Ryerson AB, Eheman CR, Altekruse SF, et al. Annual Report to the Nation on the Status of Cancer, 1975–2012, featuring the increasing incidence of liver cancer. Cancer. 2016;122(9):1312–1337. doi:10.1002/cncr.29936
30. Nguyen BN, Fléjou JF, Terris B, Belghiti J, Degott C. Focal nodular hyperplasia of the liver: a comprehensive pathologic study of 305 lesions and recognition of new histologic forms. Am J Surg Pathol. 1999;23(12):1441–1454. doi:10.1097/00000478-199912000-00001
31. McGlynn KA, London WT. The global epidemiology of hepatocellular carcinoma: present and future. Clin Liver Dis. 2011;15(2):223–243, vii–x. doi:10.1016/j.cld.2011.03.006
32. Walters GO, Miller FM, Worwood M. Serum ferritin concentration and iron stores in normal subjects. J Clin Pathol. 1973;26(10):770–772. doi:10.1136/jcp.26.10.770
33. Tran KT, Coleman HG, McCain RS, Cardwell CR. Serum biomarkers of iron status and risk of primary liver cancer: a systematic review and meta-analysis. Nutr Cancer. 2019;71(8):1365–1373. doi:10.1080/01635581.2019.1609053
34. Tang DJ, Lang YM, Feng YZ. Evaluation of combined assays of serum ferritin, alpha-1-antitrypsin and alpha-fetoprotein in liver cancer. Chinese Med J. 1992;105(11):900–904.
35. Nakano S, Kumada T, Sugiyama K, Watahiki H, Takeda I. Clinical significance of serum ferritin determination for hepatocellular carcinoma. Am J Gastroenterol. 1984;79(8):623–627.
36. Galle PR, Foerster F, Kudo M, et al. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int. 2019;39(12):2214–2229. doi:10.1111/liv.14223
37. Di Bisceglie AM, Sterling RK, Chung RT, et al. Serum alpha-fetoprotein levels in patients with advanced hepatitis C: results from the HALT-C Trial. J Hepatol. 2005;43(3):434–441. doi:10.1016/j.jhep.2005.03.019
38. Fouad R, Elsharkawy A, Abdel Alem S, et al. Clinical impact of serum α-fetoprotein and its relation on changes in liver fibrosis in hepatitis C virus patients receiving direct-acting antivirals. Eur J Gastroenterol Hepatol. 2019;31(9):1129–1134. doi:10.1097/meg.0000000000001400
39. Zhang K, Song P, Gao J, Li G, Zhao X, Zhang S. Perspectives on a combined test of multi serum biomarkers in China: towards screening for and diagnosing hepatocellular carcinoma at an earlier stage. Drug Discoveries Ther. 2014;8(3):102–109. doi:10.5582/ddt.2014.01026
40. Chen H, Zhang Y, Li S, et al. Direct comparison of five serum biomarkers in early diagnosis of hepatocellular carcinoma. Cancer Manage Res. 2018;10:1947–1958. doi:10.2147/cmar.S167036
41. de Martel C, Maucort-Boulch D, Plummer M, Franceschi S. World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology. 2015;62(4):1190–1200. doi:10.1002/hep.27969
42. Jeng WJ, Papatheodoridis GV, Lok ASF. Hepatitis B. Lancet. 2023;401(10381):1039–1052. doi:10.1016/s0140-6736(22)01468-4
43. Vo T T, Poovorawan K, Charoen P, et al. Association between hepatitis B surface antigen levels and the risk of hepatocellular carcinoma in patients with chronic hepatitis B infection: systematic review and meta-analysis. Asian Pac J Cancer Prev. 2019;20(8):2239–2246. doi:10.31557/apjcp.2019.20.8.2239
44. Tseng TC, Liu CJ, Yang HC, et al. High levels of hepatitis B surface antigen increase risk of hepatocellular carcinoma in patients with low HBV load. Gastroenterology. 2012;142(5):1140–1149.e1143; quiz e1113–1144. doi:10.1053/j.gastro.2012.02.007
45. Xi IL, Wu J, Guan J, et al. Deep learning for differentiation of benign and malignant solid liver lesions on ultrasonography. Abdom Radiol. 2021;46(2):534–543. doi:10.1007/s00261-020-02564-w
46. Yasaka K, Akai H, Abe O, Kiryu S. Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology. 2018;286(3):887–896. doi:10.1148/radiol.2017170706
47. Shen H, Lv G, Lin H, et al. Development of an ultrasound prediction model to discriminate between malignant and benign liver lesions. Ultrasound Med Biol. 2020;46(4):952–958. doi:10.1016/j.ultrasmedbio.2019.12.018
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.